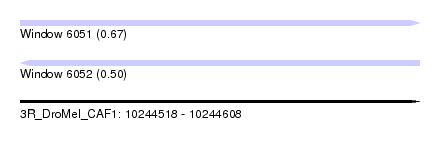
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,244,518 – 10,244,608 |
| Length | 90 |
| Max. P | 0.669356 |

| Location | 10,244,518 – 10,244,608 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.89 |
| Mean single sequence MFE | -35.73 |
| Consensus MFE | -21.71 |
| Energy contribution | -21.27 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
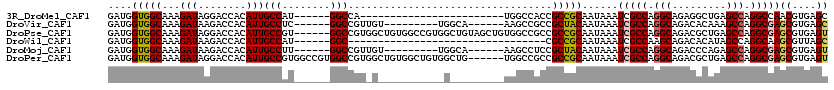
>3R_DroMel_CAF1 10244518 90 + 27905053 GCUCACGUUGGCCUGGCUCAGCCUCUGCCUGGCGAUUUAUUGCGGCGGUGGCCA------------------------UGGCC------AUGGCAAUGUGGUCCUAUCUUUGCCACCAUC ..((((((((.(((((((..(((.(((((..((((....))))))))).)))..------------------------.))))------).))))))))))................... ( -36.10) >DroVir_CAF1 118809 99 + 1 GCUCACGCUCGCCUGGCUUUGUGUCUGCCUGGCGAUUUAUUGUAGCGGCGGCUU------UGCCA---------ACAACGGCC------GAGGCAAUGUGGUCUUAUCUUUGCCACCAUC (((((...(((((.(((.........))).))))).....)).)))((.(((..------.(((.---------.....))).------(((((......)))))......))).))... ( -33.10) >DroPse_CAF1 110339 114 + 1 ACUCACGCUCGCCUGGCUCAGCGUCUGCCUGGCGAUUUAUUGCGGCGGCGGCCACAGCUACAGCCACGGCCACAGCCACGGCC------ACGGCAAUGUGGUCCUAUCUUUGCCACCAUC ......(((((((.(((.........))).)))))......))((.(((((((...((....))...))))........((((------(((....)))))))........))).))... ( -42.30) >DroWil_CAF1 115597 81 + 1 GCUAACGCUUGCCUGGCUAUGUGUCUGCUUGGCGAUUUAUUGCGGCG---------------------------------GCC------AUGGCAAUGUGGUCUUAUCUUUGCCACCAUC ((....)).......((((((.(.(((((..((((....))))))))---------------------------------)))------)))))...(((((.........))))).... ( -23.90) >DroMoj_CAF1 111372 99 + 1 ACUCACGCUCGCCUGGCUCUGGGUCUGCCUGGCGAUUUAUUGUAGCGGAGGCUU------UGCCA---------ACAACGGCC------AAGGCAAUGUGGUCUUAUCUUUGCCACCAUC .(((.((((((((.(((.........))).))))).........))))))((((------((((.---------.....)).)------)))))...(((((.........))))).... ( -34.00) >DroPer_CAF1 113024 114 + 1 ACUCACGCUCGCCUGGCUCAGCGUCUGCCUGGCGAUUUAUUGCGGCGGCGGCCA------CAGCCACAGCCACAGCCACGGCCACGGCCACGGCAAUGUGGUCCUAUCUUUGCCACCAUC ......(((((((.(((.........))).)))))......))((.((((((..------..)))...(((((((((..(((....)))..)))..)))))).........))).))... ( -45.00) >consensus ACUCACGCUCGCCUGGCUCAGCGUCUGCCUGGCGAUUUAUUGCGGCGGCGGCCA_________C___________C_ACGGCC______AAGGCAAUGUGGUCCUAUCUUUGCCACCAUC .....((((((((.(((.........))).)))).........)))).................................(((........)))...(((((.........))))).... (-21.71 = -21.27 + -0.44)

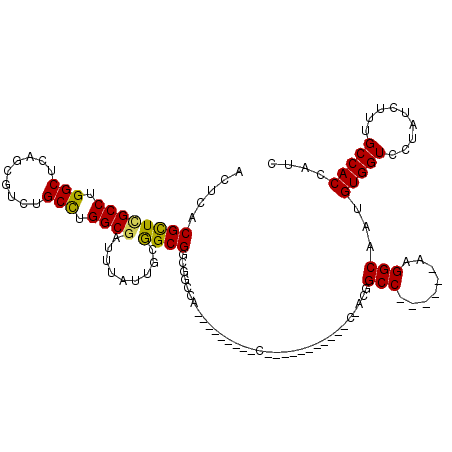
| Location | 10,244,518 – 10,244,608 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.89 |
| Mean single sequence MFE | -37.24 |
| Consensus MFE | -21.33 |
| Energy contribution | -21.38 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.57 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10244518 90 - 27905053 GAUGGUGGCAAAGAUAGGACCACAUUGCCAU------GGCCA------------------------UGGCCACCGCCGCAAUAAAUCGCCAGGCAGAGGCUGAGCCAGGCCAACGUGAGC .((((((((((.(.........).)))))))------((((.------------------------(((((((((((((........))..)))...)).)).))))))))..))).... ( -30.10) >DroVir_CAF1 118809 99 - 1 GAUGGUGGCAAAGAUAAGACCACAUUGCCUC------GGCCGUUGU---------UGGCA------AAGCCGCCGCUACAAUAAAUCGCCAGGCAGACACAAAGCCAGGCGAGCGUGAGC ..(((((((........(((.((...((...------.)).)).))---------)(((.------..)))))))))).......(((((.(((.........))).)))))((....)) ( -33.20) >DroPse_CAF1 110339 114 - 1 GAUGGUGGCAAAGAUAGGACCACAUUGCCGU------GGCCGUGGCUGUGGCCGUGGCUGUAGCUGUGGCCGCCGCCGCAAUAAAUCGCCAGGCAGACGCUGAGCCAGGCGAGCGUGAGU (.(((((((.......((.(......)))..------(((((..((((..((....))..))))..)))))))))))))......(((((.(((.........))).)))))((....)) ( -54.50) >DroWil_CAF1 115597 81 - 1 GAUGGUGGCAAAGAUAAGACCACAUUGCCAU------GGC---------------------------------CGCCGCAAUAAAUCGCCAAGCAGACACAUAGCCAGGCAAGCGUUAGC (((((((((........).)))).(((((.(------(((---------------------------------.((.((........))...)).(...)...))))))))).))))... ( -18.00) >DroMoj_CAF1 111372 99 - 1 GAUGGUGGCAAAGAUAAGACCACAUUGCCUU------GGCCGUUGU---------UGGCA------AAGCCUCCGCUACAAUAAAUCGCCAGGCAGACCCAGAGCCAGGCGAGCGUGAGU ....(((((........).)))).(((((((------(((..((((---------((...------.(((....))).))))))...))))(((.........)))))))))((....)) ( -29.40) >DroPer_CAF1 113024 114 - 1 GAUGGUGGCAAAGAUAGGACCACAUUGCCGUGGCCGUGGCCGUGGCUGUGGCUGUGGCUG------UGGCCGCCGCCGCAAUAAAUCGCCAGGCAGACGCUGAGCCAGGCGAGCGUGAGU (.(((((((.......((.(((((..((((..((((..((....))..))))..))))))------)))))))))))))......(((((.(((.........))).)))))((....)) ( -58.21) >consensus GAUGGUGGCAAAGAUAAGACCACAUUGCCAU______GGCCGU_G___________G_________UGGCCGCCGCCGCAAUAAAUCGCCAGGCAGACGCAGAGCCAGGCGAGCGUGAGC ....(((((...(((........)))(((........)))..................................)))))......(((((.(((.........))).)))))((....)) (-21.33 = -21.38 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:48 2006