
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,237,914 – 10,238,053 |
| Length | 139 |
| Max. P | 0.950234 |

| Location | 10,237,914 – 10,238,015 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 84.27 |
| Mean single sequence MFE | -39.85 |
| Consensus MFE | -33.08 |
| Energy contribution | -33.50 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
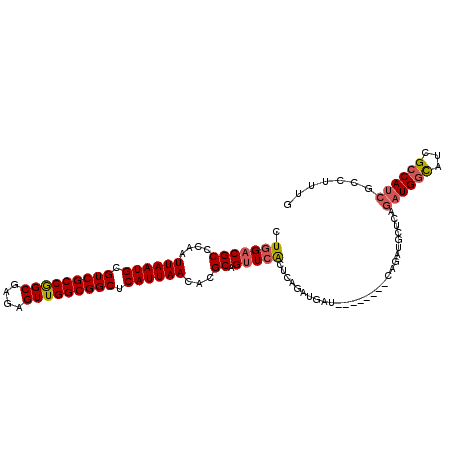
>3R_DroMel_CAF1 10237914 101 - 27905053 CUGGACGCCCAAUUAAUGCGUCGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUCACUGAGAUGAU--------CAGAUGCUCAGAUGGCAUCGCCAUCGCCUUUG .(((((((....((((((.(((((((((....))))))))).))))))...)).)))))((((.....)--------))).......((((((...))))))....... ( -40.40) >DroVir_CAF1 112807 103 - 1 AUGGCCGCCCAAUUAAUGCGUCGCCAGCGAGAGUUGGCGGCUCAUUAACACGCAGUUGGCCCGUACGCC---AUGC-CAUUGG-GUA-UUGGUAUCGGCAUCGACAAUG ..((((((....((((((.(((((((((....))))))))).))))))...))....))))((...(((---((((-((....-...-.)))))).)))..))...... ( -43.10) >DroSec_CAF1 95801 101 - 1 CUGGACGCCCAAUUAAUGCGUCGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUCACUCAGAUUAU--------CAGAUGCUCAGAUGGCAUCGCCAUCGUCUUUG .(((((((....((((((.(((((((((....))))))))).))))))...)).)))))...((((.((--------..((((((.....))))))...)).))))... ( -37.00) >DroSim_CAF1 94238 101 - 1 CUGGACGCCCAAUUAAUGCGUCGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUCACUCAGAUGAU--------CAGAUGCUCAGAUGGCAUCGCCAUCGCCUUUG .(((((((....((((((.(((((((((....))))))))).))))))...)).)))))....((((..--------..((((((.....))))))..))))....... ( -39.30) >DroEre_CAF1 96732 105 - 1 CUGGACGCCCAAUUAAUGCGUCGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUCACUCAGAUGCUCCGAUGCUCAGAUGUUCAGAUGGCAUCGCCAUCGAC---- .(((((((....((((((.(((((((((....))))))))).))))))...)).)))))....((((...((((((.((..(....)..)))))))).))))...---- ( -41.30) >DroYak_CAF1 102134 93 - 1 CUGGACGCCCAAUUAAUGCGUCGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUCACUCAGAUGCU----------------CAGAUGGCAUCGCCAUCGCCUUUG .(((((((....((((((.(((((((((....))))))))).))))))...)).)))))..((((.((.----------------..((((((...)))))))).)))) ( -38.00) >consensus CUGGACGCCCAAUUAAUGCGUCGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUCACUCAGAUGAU________CAGAUGCUCAGAUGGCAUCGCCAUCGCCUUUG .(((((((....((((((.(((((((((....))))))))).))))))...)).)))))............................((((((...))))))....... (-33.08 = -33.50 + 0.42)


| Location | 10,237,946 – 10,238,053 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.07 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -22.69 |
| Energy contribution | -22.55 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10237946 107 - 27905053 AUGUUCGGUGCCCGGCCAUGCCCAUAAACGCCA--AUGAUCUGGACGCCCAAUUAAUGCGU-CGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUCACUGAGAUGAU----- ...(((((((..((((...))).......((..--......(((....))).((((((.((-(((((((....))))))))).))))))...)).)..))))))).....----- ( -37.00) >DroVir_CAF1 112839 103 - 1 -----UCAUGUGUG--GAUGCCCAUAAAAGACGUGGCGG-AUGGCCGCCCAAUUAAUGCGU-CGCCAGCGAGAGUUGGCGGCUCAUUAACACGCAGUUGGCCCGUACGCC---AU -----...((((((--................(.(((((-....))))))..((((((.((-(((((((....))))))))).))))))))))))..((((......)))---). ( -45.80) >DroPse_CAF1 103932 92 - 1 -----CGAUGCGUAGCCAUGCCCAUAAACGACA--AUGAUCCGGACGCCCAAUUAAUGCGUCCGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUG---------------- -----...(((((.((...))...........(--((((.((((((((.........))))))((((((....)))))))).)))))...)))))....---------------- ( -33.70) >DroEre_CAF1 96763 112 - 1 AUGCCUGCUUCCCUACCAUGCCCAUAAACGCCA--AUGAUCUGGACGCCCAAUUAAUGCGU-CGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUCACUCAGAUGCUCCGAU .............................((..--.(((..(((((((....((((((.((-(((((((....))))))))).))))))...)).))))).)))...))...... ( -32.50) >DroAna_CAF1 87118 93 - 1 AUG----AUUCGCGGCCAUGCCCGUAAACGACU--AUGAUCCGGAUGCCCAAUUAAUGCGU-CGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUCG--------------- ...----(((((.(..((((..((....))..)--)))..))))))((....((((((.((-(((((((....))))))))).))))))...))......--------------- ( -33.50) >DroPer_CAF1 106559 92 - 1 -----CGAUGCGUAGCCAUGCCCAUAAACGACA--AUGAUCCGGACGCCCAAUUAAUGCGUCCGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUG---------------- -----...(((((.((...))...........(--((((.((((((((.........))))))((((((....)))))))).)))))...)))))....---------------- ( -33.70) >consensus _____CGAUGCGCGGCCAUGCCCAUAAACGACA__AUGAUCCGGACGCCCAAUUAAUGCGU_CGCCGGCGAGAGUUGGCGGCUCAUUAACACGCAGUUC________________ ..................(((.....................((....))..((((((.((.(((((((....))))))))).))))))...))).................... (-22.69 = -22.55 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:43 2006