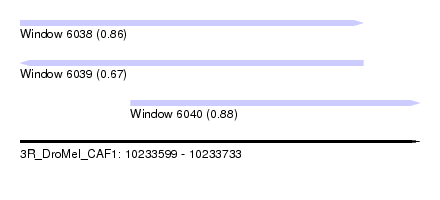
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,233,599 – 10,233,733 |
| Length | 134 |
| Max. P | 0.875106 |

| Location | 10,233,599 – 10,233,714 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.44 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -14.29 |
| Energy contribution | -15.32 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10233599 115 + 27905053 -CAGGCUC-ACAAAUAAUUAUAGAAAUUAUGAAUCAA-GCCCCAUCGGACAGACUGUUAUUAUAAACCACUCAGGCUUAAUAACAGACCGCUAAUUCGCUGGCAGGACCACCGGCGCA -..((((.-....((((((.....))))))......)-)))...((((.....(((((((((....((.....))..))))))))).((((((......)))).))....)))).... ( -25.90) >DroVir_CAF1 108889 109 + 1 -AAAUAUCUGACCAUAAUUAUAGAAAUUAUGAAACGC-AGCC-------UUCACUGUUAUUAUAAACCACUCACGCUUAAUAACAGGCCGCUAAUUGGCUGGCAGGACCACCAGCGCA -...........(((((((.....)))))))...(((-.(((-------....(((((((((...............)))))))))((((.....)))).))).((....)).))).. ( -25.36) >DroGri_CAF1 96540 110 + 1 AAAAAUUCUAGUCAUAAUUAUAGAAAUUAUGAAAUAC-AGUC-------UGCACUGUUAUUAUAAACCACUCACGCUUAAUAACAGGCCGCUAAUUGGCUGGCAGGACCACCAGCGCA ...........((((((((.....))))))))....(-(((.-------.((.(((((((((...............)))))))))...))..))))(((((........)))))... ( -26.36) >DroWil_CAF1 105601 112 + 1 -CGGUCUAUGCUAAUAAUUAUAGAAAUUAUGAAACUCAGCCAC-----ACACACUAUUAUUAUAAACCACUCAGGCUUAAUAACAGGUCGCUAAUUGGCUGGCAGGACCACCAGCACA -.(((((.(((..((((((.....))))))......((((((.-----.....((.((((((....((.....))..)))))).)).........))))))))))))))......... ( -24.06) >DroAna_CAF1 83407 111 + 1 -CAGGUCC-GAGCCUAAUUAUUGAAAUAAUGAAUCAU-GCCCCA----GUACGCUGUUAUUAUAAACCACUCAGGCUUAAUAACAGGCCGCUAAUUGGUCGGCAGGACCACCGGCGAA -..(((((-((.....(((((....)))))...)).(-((((((----((.(((((((((((....((.....))..)))))))))..))...)))))..)))))))))......... ( -29.70) >DroPer_CAF1 102858 111 + 1 -CAGACUC-UGUCAUAAUUAUAGAAAUUAUGAGUCAU-GCUUGA----CCACACUGUUAUUAUAAACCACUCAGGCUUAAUAACAGUCCACUAAUUGGCUGGCAGGACCACCAAGACC -.....((-(.((((((((.....))))))))(((.(-(((.(.----(((.((((((((((....((.....))..))))))))))........)))).)))).))).....))).. ( -25.20) >consensus _CAGACUC_GAUCAUAAUUAUAGAAAUUAUGAAACAC_GCCCC______CACACUGUUAUUAUAAACCACUCAGGCUUAAUAACAGGCCGCUAAUUGGCUGGCAGGACCACCAGCGCA ............(((((((.....)))))))......................(((((((((....((.....))..)))))))))...........(((((........)))))... (-14.29 = -15.32 + 1.03)

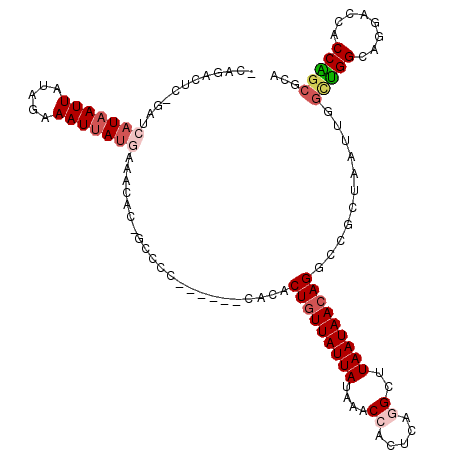
| Location | 10,233,599 – 10,233,714 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.44 |
| Mean single sequence MFE | -32.54 |
| Consensus MFE | -17.72 |
| Energy contribution | -19.00 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10233599 115 - 27905053 UGCGCCGGUGGUCCUGCCAGCGAAUUAGCGGUCUGUUAUUAAGCCUGAGUGGUUUAUAAUAACAGUCUGUCCGAUGGGGC-UUGAUUCAUAAUUUCUAUAAUUAUUUGU-GAGCCUG- ..(((.(((......))).))).....((((.(((((((((((((.....))))..))))))))).))))......((((-(((....((((((.....))))))...)-)))))).- ( -35.30) >DroVir_CAF1 108889 109 - 1 UGCGCUGGUGGUCCUGCCAGCCAAUUAGCGGCCUGUUAUUAAGCGUGAGUGGUUUAUAAUAACAGUGAA-------GGCU-GCGUUUCAUAAUUUCUAUAAUUAUGGUCAGAUAUUU- ((.((((((......))))))))....((((((((((((((.((....))......))))))).....)-------))))-))...((((((((.....))))))))..........- ( -32.90) >DroGri_CAF1 96540 110 - 1 UGCGCUGGUGGUCCUGCCAGCCAAUUAGCGGCCUGUUAUUAAGCGUGAGUGGUUUAUAAUAACAGUGCA-------GACU-GUAUUUCAUAAUUUCUAUAAUUAUGACUAGAAUUUUU ((.((((((......))))))))..((((.(((((((((((.((....))......))))))))).)).-------).))-)....((((((((.....))))))))........... ( -30.30) >DroWil_CAF1 105601 112 - 1 UGUGCUGGUGGUCCUGCCAGCCAAUUAGCGACCUGUUAUUAAGCCUGAGUGGUUUAUAAUAAUAGUGUGU-----GUGGCUGAGUUUCAUAAUUUCUAUAAUUAUUAGCAUAGACCG- ((((((((((((...((((((((....(((..(((((((((((((.....))))..)))))))))..)))-----.)))))).))...............)))))))))))).....- ( -33.97) >DroAna_CAF1 83407 111 - 1 UUCGCCGGUGGUCCUGCCGACCAAUUAGCGGCCUGUUAUUAAGCCUGAGUGGUUUAUAAUAACAGCGUAC----UGGGGC-AUGAUUCAUUAUUUCAAUAAUUAGGCUC-GGACCUG- ..(((..((((((.....)))).))..)))(((((((((((((((.....))))..))))))))).)).(----((((.(-.(((((.(((.....))))))))).)))-)).....- ( -32.10) >DroPer_CAF1 102858 111 - 1 GGUCUUGGUGGUCCUGCCAGCCAAUUAGUGGACUGUUAUUAAGCCUGAGUGGUUUAUAAUAACAGUGUGG----UCAAGC-AUGACUCAUAAUUUCUAUAAUUAUGACA-GAGUCUG- .((((((((((.....))).))))...((((((((((((((((((.....))))..)))))))))(((((----(((...-.)))).))))...)))))......))).-.......- ( -30.70) >consensus UGCGCUGGUGGUCCUGCCAGCCAAUUAGCGGCCUGUUAUUAAGCCUGAGUGGUUUAUAAUAACAGUGUA______GGGGC_GUGAUUCAUAAUUUCUAUAAUUAUGACC_GAGACUG_ ...((((((......))))))...........(((((((((((((.....))))..))))))))).....................((((((((.....))))))))........... (-17.72 = -19.00 + 1.28)


| Location | 10,233,636 – 10,233,733 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 79.75 |
| Mean single sequence MFE | -21.25 |
| Consensus MFE | -14.33 |
| Energy contribution | -14.50 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10233636 97 + 27905053 CCCA--UCGGACAGACUGUUAUUAUAAACCACUCAGGCUUAAUAACAGACCGCUAAUUCGCUGGCAGGACCACCGGCGCAUCAAAUCAGGCAUUCA-GCC ....--((((.....(((((((((....((.....))..))))))))).((((((......)))).))....))))............(((.....-))) ( -22.10) >DroVir_CAF1 108927 90 + 1 CC---------UUCACUGUUAUUAUAAACCACUCACGCUUAAUAACAGGCCGCUAAUUGGCUGGCAGGACCACCAGCGCAUCAAAUCAAC-AAGCCUCCC ((---------(...(((((((((...............)))))))))((((((....))).))))))......................-......... ( -17.06) >DroGri_CAF1 96579 89 + 1 UC---------UGCACUGUUAUUAUAAACCACUCACGCUUAAUAACAGGCCGCUAAUUGGCUGGCAGGACCACCAGCGCAUCAAAUCAAC-AUGCC-ACC ((---------(((.(((((((((...............)))))))))((((.....))))..))))).......(.((((.........-)))))-... ( -19.96) >DroYak_CAF1 97559 99 + 1 CCCACCACGGACAGACUGUUAUUAUAAACCACUCAGGCUUAAUAACAGAUCGCUAAUUGGCUGGCAGGACCACCGGCGCAUCAAAUCAGCCAUUUA-GCC .((.....))...(((((((((((....((.....))..))))))))).))(((((.((((((((.((....)).)).........)))))).)))-)). ( -29.00) >DroAna_CAF1 83444 90 + 1 CCCA------GUACGCUGUUAUUAUAAACCACUCAGGCUUAAUAACAGGCCGCUAAUUGGUCGGCAGGACCACCGGCGAAUCAAAUCAGACAUUCA---- ....------((...(((((((((....((.....))..)))))))))((((.....(((((.....))))).))))............)).....---- ( -21.80) >DroPer_CAF1 102895 90 + 1 UUGA------CCACACUGUUAUUAUAAACCACUCAGGCUUAAUAACAGUCCACUAAUUGGCUGGCAGGACCACCAAGACCUCAAAUCAGACAG----ACC ((((------....((((((((((....((.....))..)))))))))).......((((.(((.....)))))))....)))).........----... ( -17.60) >consensus CCCA_______CACACUGUUAUUAUAAACCACUCAGGCUUAAUAACAGGCCGCUAAUUGGCUGGCAGGACCACCAGCGCAUCAAAUCAGCCAUUCA_ACC ...............(((((((((....((.....))..)))))))))...........(((((........)))))....................... (-14.33 = -14.50 + 0.17)

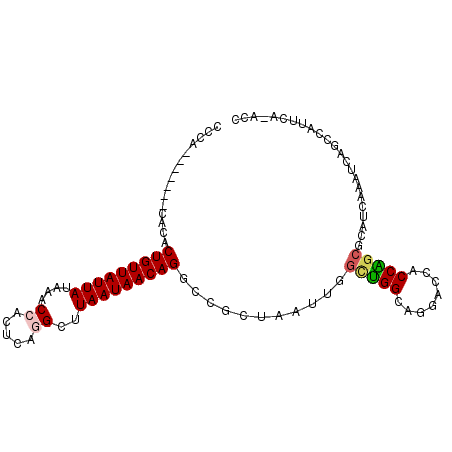
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:37 2006