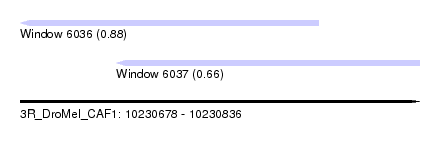
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,230,678 – 10,230,836 |
| Length | 158 |
| Max. P | 0.878290 |

| Location | 10,230,678 – 10,230,796 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.59 |
| Mean single sequence MFE | -36.10 |
| Consensus MFE | -22.61 |
| Energy contribution | -22.45 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10230678 118 - 27905053 CAGGAGCUUCUCGCAGAAGUUCGAGAACUGGCCAAAACGCAAGGCCGAGGCGUUAUGGCGGAAAAUGCGCGAACGCAAGAUUGCAGGUGAGC-AGAAAU-UGGCAUUUGCUGACCACCAA ...(((((((.....)))))))........((((.(((((.........))))).))))((.......((((.(....).)))).(((.(((-(((...-.....)))))).))).)).. ( -42.30) >DroVir_CAF1 106190 107 - 1 UAGGAGCUUCUCCCAGAAGUUCGAGAACUGGCCCAAACGCAAGGCCGAGGCGUUGUGGCGAAAAAUGCGCGAGCGCAAAAUCGCAGGUGAGC-A--AUU-UGAA-C-----GAUCGC--- ..((((...))))..((.(((((((..((.(((....(((((.((....)).)))))((((....((((....))))...)))).))).)).-.--.))-))))-)-----..))..--- ( -37.00) >DroEre_CAF1 89911 100 - 1 CAGGAGCUUCUCGCAGAAGUUCGAGAACUGGCCAAAACGCAAGGCCGAGGCGUUGUGGCGGAAAAUGCGCGAACGCAAGAUUGCAGGUGAGC-AGA--------ACUGG----------- ((((((((((.....))))))).....((.(((....(((((.((....)).)))))(((.....)))((((.(....).)))).))).)).-...--------.))).----------- ( -36.30) >DroWil_CAF1 102600 106 - 1 UAGGAGCUUCUCGCAAAAAUUCGAGAAUUGGCCCAAACGCAAGGCCGAGGCACUAUGGCGAAAAAUGCGCGAACGUAAAAUUGCAGGUAAGCCAGAAAGCUAGCAA-------------- .....((((((((........))))))((((((.........))))))(((....((((......((((............)))).....))))....))).))..-------------- ( -26.60) >DroMoj_CAF1 98283 109 - 1 CAGGAGCUUCUCCCAAAAGUUCGAGAAUUGGCCCAAGCGCAAGGCCGAGGCGCUUUGGCGCAAAAUGCGAGAGCGCAAAAUCGCAGGUGAGC-ACCACU-CGGA-U-----GAUCGA--- ..((((...)))).....(((((((..((((((.........)))))).(((((((.(((.....))).))))))).........(((....-))).))-))))-)-----......--- ( -37.40) >DroAna_CAF1 80931 112 - 1 CAGGAGCUUCUCGCAGAAGUUUGAGAACUGGCCGAAGCGCAAGGCCGAGGCGUUAUGGCGGAAGAUGCGGGAACGCAAGAUUGCAGGUGAGA-ACGGGC-UAGGAUCU-CUGAC-----A ...(((((((.....)))))))((((.((((((....(....)(((...(((......(....).((((....))))....))).)))....-...)))-)))..)))-)....-----. ( -37.00) >consensus CAGGAGCUUCUCGCAGAAGUUCGAGAACUGGCCCAAACGCAAGGCCGAGGCGUUAUGGCGGAAAAUGCGCGAACGCAAAAUUGCAGGUGAGC_AGAAAU_UGGAAU_____GA_______ ...(((((((.....))))))).....((.(((..(((((.........)))))...((((....((((....))))...)))).))).))............................. (-22.61 = -22.45 + -0.16)

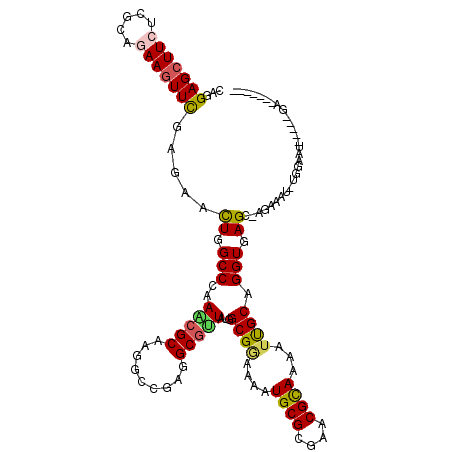
| Location | 10,230,716 – 10,230,836 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -43.72 |
| Consensus MFE | -33.69 |
| Energy contribution | -32.75 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10230716 120 - 27905053 AACGUCGCAUGUCCAUCACCAUACGCGAGUACAAGUUCUUCAGGAGCUUCUCGCAGAAGUUCGAGAACUGGCCAAAACGCAAGGCCGAGGCGUUAUGGCGGAAAAUGCGCGAACGCAAGA ..((.(((((.(((....((((((((.......(((((((...(((((((.....))))))))))))))((((.........))))...))).))))).)))..)))))))..(....). ( -43.70) >DroVir_CAF1 106217 120 - 1 AACGCCGCAUGUCUGUAUCUAUACGGGAGUACAAGUUCUUUAGGAGCUUCUCCCAGAAGUUCGAGAACUGGCCCAAACGCAAGGCCGAGGCGUUGUGGCGAAAAAUGCGCGAGCGCAAAA ..((((((((((((((((((.....))).))).(((((((...(((((((.....))))))))))))))((((.........)))).))))).))))))).....((((....))))... ( -45.80) >DroGri_CAF1 93825 120 - 1 AGCGCCGCAUGUCCGUAUCGAUACGGGAGUACAAGUUCUUUAGGAGCUUCUCCCAAAAAUUCGAGAACUGGCCAAAACGGAAGGCGGAGGCGUUGUGGCGCAAAAUGCGCGAGCGUAAAA .(((((((((.(((((.((((...(((((...((((((.....)))))))))))......))))...((..((.....)).))))))).)...))))))))...((((....)))).... ( -46.90) >DroWil_CAF1 102626 120 - 1 AACGUCGUAUGUCCGUCACGAUACGCGAGUACAAGUUCUUUAGGAGCUUCUCGCAAAAAUUCGAGAAUUGGCCCAAACGCAAGGCCGAGGCACUAUGGCGAAAAAUGCGCGAACGUAAAA ..(((((((((((..((.(((...(((((...((((((.....)))))))))))......))).)).((((((.........)))))))))).))))))).....((((....))))... ( -36.50) >DroMoj_CAF1 98312 120 - 1 AGCGCCGCAUGUCCGUAUCGAUACGGGAGUACAAGUUCUUCAGGAGCUUCUCCCAAAAGUUCGAGAAUUGGCCCAAGCGCAAGGCCGAGGCGCUUUGGCGCAAAAUGCGAGAGCGCAAAA .(((((((((.......((((...(((((...((((((.....)))))))))))......))))...((((((.((((((.........)))))).))).))).)))))...)))).... ( -44.70) >DroAna_CAF1 80963 120 - 1 AACGUCGCAUGUCCGUCACGAUUCGCGAGUACAAGUUCUUCAGGAGCUUCUCGCAGAAGUUUGAGAACUGGCCGAAGCGCAAGGCCGAGGCGUUAUGGCGGAAGAUGCGGGAACGCAAGA ....((((((.(((((((..(..(((.......(((((((...(((((((.....))))))))))))))((((.........))))...))))..)))))))..))))))...(....). ( -44.70) >consensus AACGCCGCAUGUCCGUAACGAUACGCGAGUACAAGUUCUUCAGGAGCUUCUCCCAAAAGUUCGAGAACUGGCCCAAACGCAAGGCCGAGGCGUUAUGGCGAAAAAUGCGCGAACGCAAAA ..(((((..((((...........(((((...((((((.....)))))))))))..............(((((.........))))).))))...))))).....((((....))))... (-33.69 = -32.75 + -0.94)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:34 2006