| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,206,796 – 10,206,916 |
| Length | 120 |
| Max. P | 0.919447 |

| Location | 10,206,796 – 10,206,916 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.61 |
| Mean single sequence MFE | -42.88 |
| Consensus MFE | -19.32 |
| Energy contribution | -21.13 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.34 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
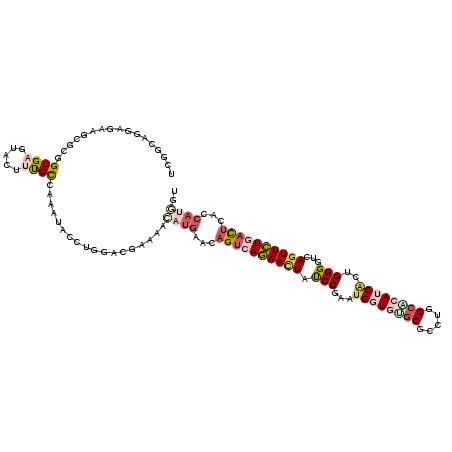
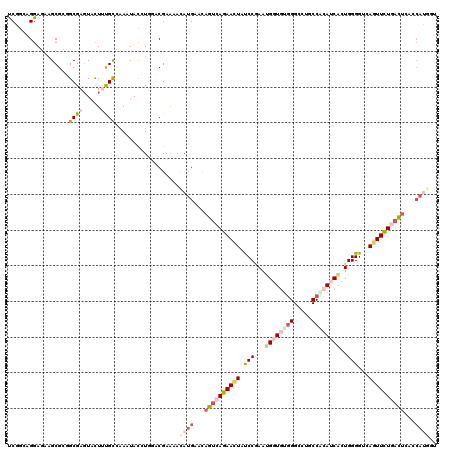
>3R_DroMel_CAF1 10206796 120 - 27905053 UCGCGAGGAUCAGCGAGGCGAGUACUUUGCAAAUUACCUCGUCGAGAAUAUGACCAGUCAGAACUACCCGAAUGGUAUGGGUCUGCCACAGCAUUGGGGUCAGUUCUGAUUCGCCAUAGU ((((((((....((((((......))))))......))))).)))...((((.(.((((((((((((((.((((.(.(((.....))).).)))).)))).)))))))))).).)))).. ( -47.60) >DroPse_CAF1 72340 112 - 1 UCCGCCGGAGAAGCGCUGCGAGUACUAUGUCAAAUACAUGGACGAACACAUGAACAGUCAGAAUUAUCCGAAUGGUAUGGGCCUGCCCCAUCGCUGGGGCCAGUUCUGACUG-------- ..(((((......))..))).((.((((((.....))))))))...........(((((((((((........(((....))).((((((....)))))).)))))))))))-------- ( -36.00) >DroEre_CAF1 65405 120 - 1 UCGGGAGGAUCAGCGAGGCGAGUACUUUGCAAAUUACCUGGUCGAGAACAUGACCAGUCAGAACUAUCCGAAUGGUGUGGGUCUGCCACAGCAUUGGGGUGAGUUCUGAUUCGCCAUAGU (((.(.....)..)))(((((((..............(((((((......))))))).(((((((((((.((((.(((((.....))))).)))).)))).))))))))))))))..... ( -49.30) >DroWil_CAF1 75024 119 - 1 CCAACCGGAGAUGCGUCGCGAUUAUUUUGCCAACUAUUUGUACGAGAACAUGAAUAGCAAAAACUAUCCAAAUGGUGAGGGCUUGCCUCAUCAUUGGGGACAAUUUUGAC-CACCAUGCA ......((...((.((((.((((.((((((....((((..(........)..))))))))))....(((.((((((((((.....))))))))))..))).)))).))))-))))..... ( -29.40) >DroAna_CAF1 58131 120 - 1 CCGGGAGGAGCAGCGCGGCGAGUUCUUCGCCGAGUACCUCUUCGAAAACAUGACCAGUGAGAACUAUCCCAACGGUGUUGGCCUGCCGCAUCACUGGGGUGAGUUCUGAUUGAGCAUGGG ((..((((((..((.(((((((...))))))).))..)))))).....((((..((((.((((((((((((..(((((.((....)))))))..)))))).)))))).))))..)))))) ( -53.70) >DroPer_CAF1 73171 120 - 1 UCCGCCGGAGAAGCGCUGCGAGUACUAUGUCAAAUAUAUGGACGAACACAUGAACAGUCAGAACUAUCCGAAUGGUGUGGGCCUGCCCCAUCGCUGGGGCCAGUUCUGACUGCACAGGAU (((..((.((.....)).)).((.((((((.....)))))))).......((..(((((((((((.(((.(.(((((.(((....)))))))).).)))..)))))))))))..))))). ( -41.30) >consensus UCGGCAGGAGAAGCGCGGCGAGUACUUUGCCAAAUACCUGGACGAAAACAUGAACAGUCAGAACUAUCCGAAUGGUGUGGGCCUGCCACAUCACUGGGGUCAGUUCUGACUCACCAUGGU .................((((.....))))..................((((...((((((((((.(((...((((((((.....))))))))..)))...))))))))))...)))).. (-19.32 = -21.13 + 1.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:27 2006