
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,170,556 – 10,170,671 |
| Length | 115 |
| Max. P | 0.505602 |

| Location | 10,170,556 – 10,170,671 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.60 |
| Mean single sequence MFE | -35.78 |
| Consensus MFE | -24.30 |
| Energy contribution | -23.55 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.505602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
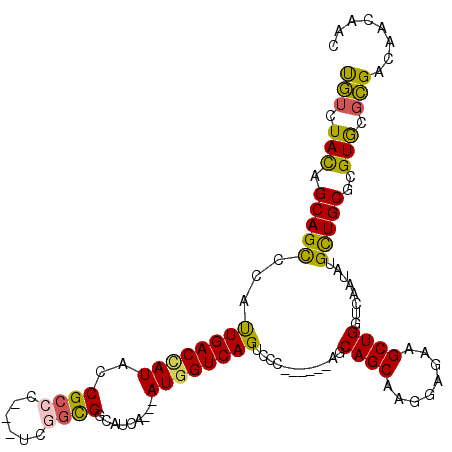
>3R_DroMel_CAF1 10170556 115 - 27905053 CAACUAUAGCAGUCCGCUGACCAUACCGCCCACCUCGAUGACCACGCACAUGGUCAGUCCCAUCGGGAACAGCAAGGAGAAGCUGGUGAAUAUGUUGCGCGUGCGUGACAACAAC ((....((((..(((((((...................((((((......)))))).((((...)))).))))..)))...)))).))....(((..((....))..)))..... ( -30.90) >DroVir_CAF1 29775 103 - 1 UGUCUACAGCAGCUCAUUGACUAUACCGCCC---UCGGCGGCAUCA---AUGGUCAGUCCC------AGCAGCAAGGAGAAGCUGGUUAAUAUGCUGCGCGUGCGCGAUAACAAC ((((....(((((..(((((((((.(((((.---..))))).....---))))))))).((------(((..(.....)..))))).......)))))((....))))))..... ( -35.40) >DroPse_CAF1 32419 106 - 1 UGUGUACAGCAGUCCUUUGACCAUACCGCCCACAUCCGUGUCAUCG---AUGGUCAGUCCG------AGCAGCAAGGAGAAGCUGGUCAACAUGCUGCGCGUGCGCGACAACAAC .((((((.(((((...((((((((......(((....)))......---))))))))...(------(.((((........)))).)).....)))))..))))))......... ( -36.90) >DroGri_CAF1 32210 103 - 1 UGUCUACAGCAGCUCAUUGACCAUACCGCCA---UCGGCGGCCUCA---AUGGUCAGUCCC------AGCAGCAAGGAGAAGCUGGUGAAUAUGCUGCGUGUACGCGAUAACAAC .((.(((((((((..(((((((((.(((((.---..))))).....---)))))))))..(------(.((((........)))).)).....))))).)))).))......... ( -39.10) >DroMoj_CAF1 33406 103 - 1 UGUCUACAGCAGCUCAUUGACAAUACCGCCC---UCGGCGGCGUCA---AUGGUCAGCCCC------AGCAGCAAGGAGAAGCUGGUCAAUAUGCUGCGAGUGCGCGAUAAUAAC .((.(((.(((((((((((((....(((((.---..))))).))))---))))......((------(((..(.....)..))))).......)))))..))).))......... ( -35.50) >DroPer_CAF1 32656 106 - 1 UGUGUACAGCAGUCCUUUGACCAUACCGCCCACAUCCGUGUCAUCG---AUGGUCAGUCCG------AGCAGCAAGGAGAAGCUGGUCAACAUGCUGCGCGUGCGCGACAACAAC .((((((.(((((...((((((((......(((....)))......---))))))))...(------(.((((........)))).)).....)))))..))))))......... ( -36.90) >consensus UGUCUACAGCAGCCCAUUGACCAUACCGCCC___UCGGCGGCAUCA___AUGGUCAGUCCC______AGCAGCAAGGAGAAGCUGGUCAAUAUGCUGCGCGUGCGCGACAACAAC (((.(((.(((((...((((((((..((((......)))).........))))))))............((((........))))........)))))..))).)))........ (-24.30 = -23.55 + -0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:08 2006