
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,169,570 – 10,169,707 |
| Length | 137 |
| Max. P | 0.893243 |

| Location | 10,169,570 – 10,169,683 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 87.10 |
| Mean single sequence MFE | -45.98 |
| Consensus MFE | -38.04 |
| Energy contribution | -38.27 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
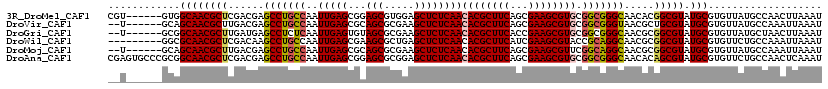
>3R_DroMel_CAF1 10169570 113 - 27905053 CGU------GUGGCAACGCUCGACGAGCCUGCCAAUUGAGCGGAGCGUGGAGCUCUCAACACGCUUCAGCGAAGCGUGCGGCGGGCAACACGGCGUAUGCGUGUUAUGCCAACUUAAAU .((------.(((((..((((.(((..((.(((....).))))..))).))))....(((((((....))...(((((((.(((....).)).)))))))))))).)))))))...... ( -49.10) >DroVir_CAF1 28781 111 - 1 --U------GCAGCAACGCUUGACGAGCCUGCCAAUUGAGCGCAGCGCGAAGCUCUCAACACGCUUCAGCGAAGCGUGCGGCGGGUAACGCUGCGUAUGCGUGUUAUGCCAAAUUAAAU --.------(((((..((.....)).(((((((..(((((.((........)).)))))((((((((...)))))))).)))))))...)))))(((((.....))))).......... ( -42.60) >DroGri_CAF1 31217 111 - 1 --U------GCGGCAACGCUUGAUGAGCCUCUCAAUUGAGUGUAGCGCGAAGCUCUCAACACGCUUCACCGAAGCGUGCGGCGGGCAACGCGGCGUAUGCGUGUUAUGCUAACUUAAAU --.------(((....)))....((((...)))).((((((.(((((((..((((.(..((((((((...))))))))..).))))..))).(((....))).....)))))))))).. ( -43.80) >DroWil_CAF1 33250 110 - 1 ---------GGCGCAACGCUCGACAAGCCUGCCAAUUGAGCGAAGCGCUGAGCUCUCAACACGCUUCAUCGAAGCGUACCGCAGGCAACGCGGCGUAUGCGUGUUCUGCCAAAUUAAAU ---------.(((((((((.((....((((((...(((((...(((.....)))))))).(((((((...)))))))...))))))....)))))).)))))................. ( -42.00) >DroMoj_CAF1 32354 111 - 1 --U------GCAGCAACGCUUGACGAGCCUGCCAAUUGAGCGCAGCGCGAAGCUCUCAACACGCUUCAGCGAAGCGUUCGGCAGGCAACGCGGCGUAUGCGUGUUAUGCCAAAUUAAAU --.------(((((((((((.(.((.(((((((....((((((..((((((((.........))))).)))..)))))))))))))..)))))))).))).)))............... ( -45.60) >DroAna_CAF1 25966 119 - 1 CGAGUGCCCGCGGCAACGCUCGACGAGCCUGCCAAUUGAGCGGAGCGCGGAGCUCUCAACACGCUUCAGCGAAGCGUGCGGCGGGCAACACAGCGUAUGCGUGUUCUGCCAACUCAAAU .(((.(((((((.(..(((((((...(....)...)))))))..)))))).)).)))..((((((((...)))))))).((((((..((((.((....))))))))))))......... ( -52.80) >consensus __U______GCGGCAACGCUCGACGAGCCUGCCAAUUGAGCGCAGCGCGAAGCUCUCAACACGCUUCAGCGAAGCGUGCGGCGGGCAACGCGGCGUAUGCGUGUUAUGCCAAAUUAAAU ............((((((((......(((((((..(((((...(((.....))))))))((((((((...)))))))).))))))).....))))).)))................... (-38.04 = -38.27 + 0.22)


| Location | 10,169,609 – 10,169,707 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 76.74 |
| Mean single sequence MFE | -34.03 |
| Consensus MFE | -23.78 |
| Energy contribution | -24.08 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10169609 98 + 27905053 CGCACGCUUCGCUGAAGCGUGUUGAGAGCUCCACGCUCCGCUCAAUUGGCAGGCUCGUCGAGCGUUGCCACACGGCACACACAUCCUACC-----UAUG-----CUCG .(((((((((...))))))))).(((.((((.(((..(((((.....))).))..))).))))((((((....)))).))..........-----....-----))). ( -35.10) >DroVir_CAF1 28820 84 + 1 CGCACGCUUCGCUGAAGCGUGUUGAGAGCUUCGCGCUGCGCUCAAUUGGCAGGCUCGUCAAGCGUUGCUGCA-------UAAA------------UUUG-----CUCG .(((((((((...)))))))))...((((...((((.(((((....((((......))))))))).)).)).-------....------------...)-----))). ( -29.90) >DroPse_CAF1 31380 101 + 1 CGCACGCUUCGCUGAAGCGUGUUGAGAGCUCUGCGCUCCGCUCAAUUGGCAGGCUCGUCGAGCGUUGCCACA-------GAUAUGUUGCCACGAGUAUGGCAGCCACG .(((((((((...)))))))))(((((((.....))))...)))..((((((((((...)))).))))))..-------.....(((((((......))))))).... ( -38.90) >DroWil_CAF1 33289 87 + 1 GGUACGCUUCGAUGAAGCGUGUUGAGAGCUCAGCGCUUCGCUCAAUUGGCAGGCUUGUCGAGCGUUGCGCC------UAAA-AUU----C-----UCAA-----CUUG ...(((((((...)))))))((((((((....((((..(((((.((.((....)).)).)))))..)))).------....-.))----)-----))))-----)... ( -34.20) >DroMoj_CAF1 32393 84 + 1 CGAACGCUUCGCUGAAGCGUGUUGAGAGCUUCGCGCUGCGCUCAAUUGGCAGGCUCGUCAAGCGUUGCUGCA-------UAAA------------UUUG-----CUCG .((.(((..(((.(((((.........))))))))..))).))....((((((((.....))).)))))(((-------....------------..))-----)... ( -27.20) >DroPer_CAF1 31606 101 + 1 CGCACGCUUCGCUGAAGCGUGUUGAGAGCUCUGCGCUCCGCUCAAUUGGCAGGCUCGUCGAGCGUUGCCACA-------GAUAUGUUGCCACGAGUAUGGCAGCCACG .(((((((((...)))))))))(((((((.....))))...)))..((((((((((...)))).))))))..-------.....(((((((......))))))).... ( -38.90) >consensus CGCACGCUUCGCUGAAGCGUGUUGAGAGCUCCGCGCUCCGCUCAAUUGGCAGGCUCGUCGAGCGUUGCCACA_______GAAAU_____C_____UAUG_____CUCG .(((((((((...)))))))))...((((..........))))...((((((((((...)))).))))))...................................... (-23.78 = -24.08 + 0.31)


| Location | 10,169,609 – 10,169,707 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 76.74 |
| Mean single sequence MFE | -34.63 |
| Consensus MFE | -20.50 |
| Energy contribution | -20.62 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
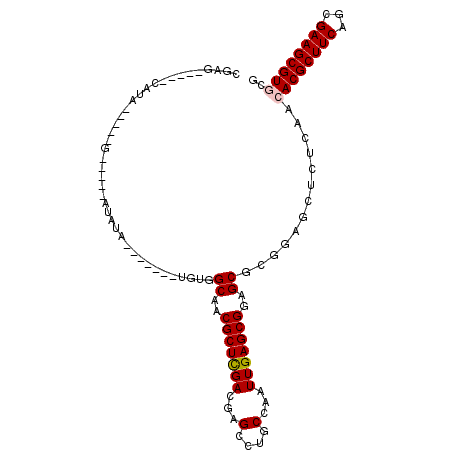
>3R_DroMel_CAF1 10169609 98 - 27905053 CGAG-----CAUA-----GGUAGGAUGUGUGUGCCGUGUGGCAACGCUCGACGAGCCUGCCAAUUGAGCGGAGCGUGGAGCUCUCAACACGCUUCAGCGAAGCGUGCG ...(-----(((.-----((((((.((.(((((((....))).)))).)).....))))))......((((((((((..........)))))))).)).....)))). ( -38.30) >DroVir_CAF1 28820 84 - 1 CGAG-----CAAA------------UUUA-------UGCAGCAACGCUUGACGAGCCUGCCAAUUGAGCGCAGCGCGAAGCUCUCAACACGCUUCAGCGAAGCGUGCG ...(-----((..------------....-------))).(((.(((((..(..(((((((......).)))).))(((((.........))))).)..)))))))). ( -26.80) >DroPse_CAF1 31380 101 - 1 CGUGGCUGCCAUACUCGUGGCAACAUAUC-------UGUGGCAACGCUCGACGAGCCUGCCAAUUGAGCGGAGCGCAGAGCUCUCAACACGCUUCAGCGAAGCGUGCG ..(((((((((((...(((....)))...-------)))))))..((((...))))..)))).......(((((.....)))))...((((((((...)))))))).. ( -39.50) >DroWil_CAF1 33289 87 - 1 CAAG-----UUGA-----G----AAU-UUUA------GGCGCAACGCUCGACAAGCCUGCCAAUUGAGCGAAGCGCUGAGCUCUCAACACGCUUCAUCGAAGCGUACC ...(-----((((-----(----(.(-((..------(((((..(((((((...(....)...)))))))..)))))))).)))))))(((((((...)))))))... ( -36.50) >DroMoj_CAF1 32393 84 - 1 CGAG-----CAAA------------UUUA-------UGCAGCAACGCUUGACGAGCCUGCCAAUUGAGCGCAGCGCGAAGCUCUCAACACGCUUCAGCGAAGCGUUCG ...(-----((..------------....-------))).(((..((((...)))).))).....((((((..((((((((.........))))).)))..)))))). ( -27.20) >DroPer_CAF1 31606 101 - 1 CGUGGCUGCCAUACUCGUGGCAACAUAUC-------UGUGGCAACGCUCGACGAGCCUGCCAAUUGAGCGGAGCGCAGAGCUCUCAACACGCUUCAGCGAAGCGUGCG ..(((((((((((...(((....)))...-------)))))))..((((...))))..)))).......(((((.....)))))...((((((((...)))))))).. ( -39.50) >consensus CGAG_____CAUA_____G_____AUAUA_______UGUGGCAACGCUCGACGAGCCUGCCAAUUGAGCGGAGCGCGGAGCUCUCAACACGCUUCAGCGAAGCGUGCG ........................................((..(((((((...(....)...)))))))..)).............((((((((...)))))))).. (-20.50 = -20.62 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:07 2006