
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,159,947 – 10,160,042 |
| Length | 95 |
| Max. P | 0.842975 |

| Location | 10,159,947 – 10,160,042 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 76.41 |
| Mean single sequence MFE | -20.37 |
| Consensus MFE | -15.95 |
| Energy contribution | -15.73 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10159947 95 + 27905053 AA----------UGGAUAUGGAAUGGAAUAAAUUUAUAAAUCAAGCACUCAUAUUAAAUCACAGUGACUAGCUAACAUGCUGGUUAGCAUCUCCUUUUUGAAAGG ..----------.(((.(((((((.......)))).............................((((((((......)))))))).))).)))........... ( -14.90) >DroSec_CAF1 18491 91 + 1 AAUGGAAUUAUUUGGAUAUGGAAUGGACUUGAUUUAUAAAUU--------------AAUCACAGUGACUAGCUAACAUGCUGGUUAGCAUCUCCGUUUUGGAAGC ..........((..((.((((((((.((((((((........--------------))))).)))(((((((......)))))))..))).)))))))..))... ( -21.70) >DroSim_CAF1 17575 91 + 1 UAUUGAAUUAUUUGGAUAUGGAAUGGACUUAAUUUAUAAAUU--------------AGUCACAGUGACUAGCUAACAUGCUGGUCAGCAUCUCCGUUUUGGACGC ..........((..((.((((((((((((.((((....))))--------------))))....((((((((......)))))))).))).)))))))..))... ( -24.50) >consensus AAU_GAAUUAUUUGGAUAUGGAAUGGACUUAAUUUAUAAAUU______________AAUCACAGUGACUAGCUAACAUGCUGGUUAGCAUCUCCGUUUUGGAAGC .............(((.(((((((.......)))).............................((((((((......)))))))).))).)))........... (-15.95 = -15.73 + -0.22)



| Location | 10,159,947 – 10,160,042 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 76.41 |
| Mean single sequence MFE | -14.70 |
| Consensus MFE | -12.34 |
| Energy contribution | -11.90 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -0.76 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10159947 95 - 27905053 CCUUUCAAAAAGGAGAUGCUAACCAGCAUGUUAGCUAGUCACUGUGAUUUAAUAUGAGUGCUUGAUUUAUAAAUUUAUUCCAUUCCAUAUCCA----------UU ...........((((((((((((......))))))..)))..((..((....(((((((.....))))))).....))..)).......))).----------.. ( -13.80) >DroSec_CAF1 18491 91 - 1 GCUUCCAAAACGGAGAUGCUAACCAGCAUGUUAGCUAGUCACUGUGAUU--------------AAUUUAUAAAUCAAGUCCAUUCCAUAUCCAAAUAAUUCCAUU ...........((((((((((((......))))))..)))(((.(((((--------------........))))))))..........)))............. ( -14.10) >DroSim_CAF1 17575 91 - 1 GCGUCCAAAACGGAGAUGCUGACCAGCAUGUUAGCUAGUCACUGUGACU--------------AAUUUAUAAAUUAAGUCCAUUCCAUAUCCAAAUAAUUCAAUA ((((((......).)))))((((.(((......))).)))).((.((((--------------((((....)))).))))))....................... ( -16.20) >consensus GCUUCCAAAACGGAGAUGCUAACCAGCAUGUUAGCUAGUCACUGUGAUU______________AAUUUAUAAAUUAAGUCCAUUCCAUAUCCAAAUAAUUC_AUU ...........((((..((((((......)))))).((((.....)))).................................))))................... (-12.34 = -11.90 + -0.44)

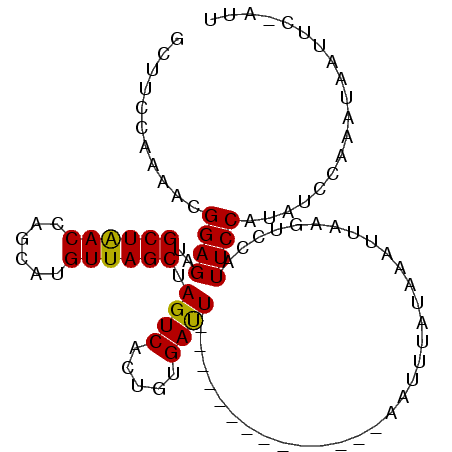

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:00 2006