
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,146,738 – 10,146,870 |
| Length | 132 |
| Max. P | 0.825963 |

| Location | 10,146,738 – 10,146,853 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 86.36 |
| Mean single sequence MFE | -21.48 |
| Consensus MFE | -15.24 |
| Energy contribution | -15.17 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10146738 115 + 27905053 UUUUUCCUUACUUGGGAUACUUCCGAAAUCCUUGUUAUUAAUUUGUAUCUUUUCUUUUUUGGUCAUCUAUGAUAAAU-CUUAUGUUGUUAUAUUGAUAUGACCGCAUAGAGAUAUU ...........((.(((....))).)).................((((((((.......(((((((.((((((((..-......)))))))).....)))))))...)))))))). ( -18.10) >DroSec_CAF1 5391 114 + 1 UUUCUCUUCACUUGGGAUACUUCCGAAAUCCUUGUUUUUAAUUCGUAUCUUUUGU--UUUGGUCAUCUAUGAUGAUUCCUUAUGUUGUUACAUUGAUAUGACCGCAUAGAGAUAUU ..(((((..((..(((((((.....(((........))).....)))))))..))--...(((((((.(((.((((..(....)..))))))).))..)))))....))))).... ( -22.00) >DroSim_CAF1 4454 114 + 1 UUUCUCUUCACUUGGGAUACUUCCGAAAUCCUUGUUUUUAAUUUGUAUCUUUUGU--UUUGGUCAUCUAUGAUGAGUCCUUAUGUUGUUAUAUUGAUAUGACCGCAUAGAGAUAUU ..((((...((..(((((((.....(((........))).....)))))))..))--...(((((((...)))))..)).(((((.((((((....)))))).))))))))).... ( -22.40) >DroYak_CAF1 4472 112 + 1 UUUUAAUUCAUUGGGGAUAUUUCUGAAAUCUGUUUUGUUUUAUUGUAUCUGUUGU--UUUGGUCAUCUAGGAUAAAU-CUUAUGUAGUUAU-UUGAUAUGACCGCAUACAGAUAUU ...(((..((..(.((((.........)))).)..))..)))..(((((((((((--...((((((...((((((..-(....)...))))-))...))))))))).)))))))). ( -23.40) >consensus UUUCUCUUCACUUGGGAUACUUCCGAAAUCCUUGUUUUUAAUUUGUAUCUUUUGU__UUUGGUCAUCUAUGAUAAAU_CUUAUGUUGUUAUAUUGAUAUGACCGCAUAGAGAUAUU ..............(((....)))....................(((((((..........(((......))).......(((((.((((((....)))))).)))))))))))). (-15.24 = -15.17 + -0.06)



| Location | 10,146,738 – 10,146,853 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 86.36 |
| Mean single sequence MFE | -17.43 |
| Consensus MFE | -10.73 |
| Energy contribution | -11.48 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10146738 115 - 27905053 AAUAUCUCUAUGCGGUCAUAUCAAUAUAACAACAUAAG-AUUUAUCAUAGAUGACCAAAAAAGAAAAGAUACAAAUUAAUAACAAGGAUUUCGGAAGUAUCCCAAGUAAGGAAAAA ......(((.(((((((((......((((.........-..)))).....)))))).............................((((..(....).))))...))).))).... ( -16.00) >DroSec_CAF1 5391 114 - 1 AAUAUCUCUAUGCGGUCAUAUCAAUGUAACAACAUAAGGAAUCAUCAUAGAUGACCAAA--ACAAAAGAUACGAAUUAAAAACAAGGAUUUCGGAAGUAUCCCAAGUGAAGAGAAA ....(((((.(((..((.((((.((((....))))..((..(((((...)))))))...--......)))).))...........((((..(....).))))...))).))))).. ( -21.70) >DroSim_CAF1 4454 114 - 1 AAUAUCUCUAUGCGGUCAUAUCAAUAUAACAACAUAAGGACUCAUCAUAGAUGACCAAA--ACAAAAGAUACAAAUUAAAAACAAGGAUUUCGGAAGUAUCCCAAGUGAAGAGAAA ....(((((.(((((((((.............(....)............))))))...--........................((((..(....).))))...))).))))).. ( -19.31) >DroYak_CAF1 4472 112 - 1 AAUAUCUGUAUGCGGUCAUAUCAA-AUAACUACAUAAG-AUUUAUCCUAGAUGACCAAA--ACAACAGAUACAAUAAAACAAAACAGAUUUCAGAAAUAUCCCCAAUGAAUUAAAA ..(((((((.((.((((((.....-((((((.....))-..)))).....))))))...--.)))))))))..................((((.............))))...... ( -12.72) >consensus AAUAUCUCUAUGCGGUCAUAUCAAUAUAACAACAUAAG_AUUCAUCAUAGAUGACCAAA__ACAAAAGAUACAAAUUAAAAACAAGGAUUUCGGAAGUAUCCCAAGUGAAGAAAAA ....(((((.(((((((((...............................)))))).............................((((..(....).))))...))).))))).. (-10.73 = -11.48 + 0.75)

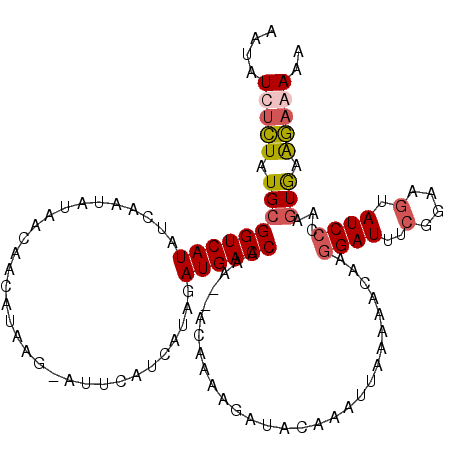

| Location | 10,146,778 – 10,146,870 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 88.93 |
| Mean single sequence MFE | -21.62 |
| Consensus MFE | -15.52 |
| Energy contribution | -15.90 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10146778 92 + 27905053 AUUUGUAUCUUUUCUUUUUUGGUCAUCUAUGAUAAAU-CUUAUGUUGUUAUAUUGAUAUGACCGCAUAGAGAUAUUGCAUGCUAUCGGAUGCA ....((((((((.......(((((((.((((((((..-......)))))))).....)))))))...))))))))(((((.(....).))))) ( -19.80) >DroSec_CAF1 5431 91 + 1 AUUCGUAUCUUUUGU--UUUGGUCAUCUAUGAUGAUUCCUUAUGUUGUUACAUUGAUAUGACCGCAUAGAGAUAUUGCAUGCUAACAGAUGCA ....(((((((((((--...(((((((.(((.((((..(....)..))))))).))..)))))))).))))))))(((((........))))) ( -20.80) >DroSim_CAF1 4494 91 + 1 AUUUGUAUCUUUUGU--UUUGGUCAUCUAUGAUGAGUCCUUAUGUUGUUAUAUUGAUAUGACCGCAUAGAGAUAUUGCAUGCUAACAGAUGCA ...(((((((.(((.--..((.(((((...)))))(((.((((((.((((((....)))))).)))))).)))....))...))).))))))) ( -21.50) >DroYak_CAF1 4512 89 + 1 UAUUGUAUCUGUUGU--UUUGGUCAUCUAGGAUAAAU-CUUAUGUAGUUAU-UUGAUAUGACCGCAUACAGAUAUUGCAUGCAAACAGCUGCA ....(((((((((((--...((((((...((((((..-(....)...))))-))...))))))))).))))))))((((.((.....)))))) ( -24.40) >consensus AUUUGUAUCUUUUGU__UUUGGUCAUCUAUGAUAAAU_CUUAUGUUGUUAUAUUGAUAUGACCGCAUAGAGAUAUUGCAUGCUAACAGAUGCA ....(((((((..........(((......))).......(((((.((((((....)))))).))))))))))))(((((........))))) (-15.52 = -15.90 + 0.38)


| Location | 10,146,778 – 10,146,870 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 88.93 |
| Mean single sequence MFE | -15.02 |
| Consensus MFE | -10.24 |
| Energy contribution | -10.30 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10146778 92 - 27905053 UGCAUCCGAUAGCAUGCAAUAUCUCUAUGCGGUCAUAUCAAUAUAACAACAUAAG-AUUUAUCAUAGAUGACCAAAAAAGAAAAGAUACAAAU .((((..((((........))))...))))((((((......((((.........-..)))).....)))))).................... ( -12.20) >DroSec_CAF1 5431 91 - 1 UGCAUCUGUUAGCAUGCAAUAUCUCUAUGCGGUCAUAUCAAUGUAACAACAUAAGGAAUCAUCAUAGAUGACCAAA--ACAAAAGAUACGAAU .(.((((....(((((.........)))))((((((....((((....))))...((....))....))))))...--.....)))).).... ( -14.10) >DroSim_CAF1 4494 91 - 1 UGCAUCUGUUAGCAUGCAAUAUCUCUAUGCGGUCAUAUCAAUAUAACAACAUAAGGACUCAUCAUAGAUGACCAAA--ACAAAAGAUACAAAU ((((((.....).))))).(((((.((((..((.(((....))).))..)))).((..(((((...)))))))...--.....)))))..... ( -13.60) >DroYak_CAF1 4512 89 - 1 UGCAGCUGUUUGCAUGCAAUAUCUGUAUGCGGUCAUAUCAA-AUAACUACAUAAG-AUUUAUCCUAGAUGACCAAA--ACAACAGAUACAAUA ((...(((((.(((((((.....)))))))((((((.....-((((((.....))-..)))).....))))))...--..)))))...))... ( -20.20) >consensus UGCAUCUGUUAGCAUGCAAUAUCUCUAUGCGGUCAUAUCAAUAUAACAACAUAAG_AUUCAUCAUAGAUGACCAAA__ACAAAAGAUACAAAU (((((........))))).(((((.((((..((.(((....))).))..)))).....(((((...)))))............)))))..... (-10.24 = -10.30 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:52 2006