| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,122,054 – 10,122,155 |
| Length | 101 |
| Max. P | 0.768249 |

| Location | 10,122,054 – 10,122,155 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.96 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -20.67 |
| Energy contribution | -21.15 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10122054 101 + 27905053 AAUUUAUGAGUGACUGAACC--GCAUUAGCCGCAUAAUUUAUGGCUCUGAUUAUGGGCCAUUCG-GCCAAUG----------------AAAAGUGGACCAACGUGGCUCAAAGUCUUGAC ...........((((.....--.....((((((.......(((((((.......)))))))..(-((((.(.----------------...).))).))...))))))...))))..... ( -27.22) >DroSec_CAF1 31320 102 + 1 AAUUUAUGAGUGACUGAGCC--GCAUUAGCCGCAUAAUUUAUGGCUCUGAUUAUGGGCCAUUCGGGCCCACG----------------AAAAGUGGACCAACGUGGCUCAAAGUCUUGAC ...........(((((((((--((....(((.(((((((.........)))))))))).....((..((((.----------------....)))).))...)))))))..))))..... ( -35.40) >DroSim_CAF1 27729 102 + 1 AAUUUAUGAGUGACUGAGCC--GCAUUAGCCGCAUAAUUUAUGGCUCUGAUUAUGGGCCAUUCGGGCCAACG----------------AAAAGUGGACCAACUUGGCUCAAAGUCUUGAC ...........(((((((((--(..((.(((((........((((((.......))))))((((......))----------------))..)))).).))..))))))..))))..... ( -31.10) >DroEre_CAF1 31064 101 + 1 AAUUUAUGAGUGCCUGAGCC--ACAUUAGCCGCAUAAUUUAUGGCUCUGAUUAUGGGCCAAUCG-GCCAACG----------------AGAAGUGGACUAACGUGGCUCCGAGUCUUGAC .......(((.(((.(((((--((.((((((((........((((((.......)))))).(((-.....))----------------)...)))).)))).))))))).).)))))... ( -37.20) >DroYak_CAF1 31820 119 + 1 AAUUUAUUAUUGCCUGAGCCGCACAUUAGCCGCAUAAUUUAUGGCUCUGAUUAUGGGCCAUUCG-GCCAACGAAAUAACGACAAAACGAAAAGCUGACCAACGUGGCUCAAAGUCUUGAC ......(((..((.((((((((...(((((...........((((((.......))))))((((-.....))))..................))))).....))))))))..))..))). ( -30.00) >consensus AAUUUAUGAGUGACUGAGCC__GCAUUAGCCGCAUAAUUUAUGGCUCUGAUUAUGGGCCAUUCG_GCCAACG________________AAAAGUGGACCAACGUGGCUCAAAGUCUUGAC ...........(((((((((.........((((........((((((.......))))))..((......))....................))))........)))))..))))..... (-20.67 = -21.15 + 0.48)


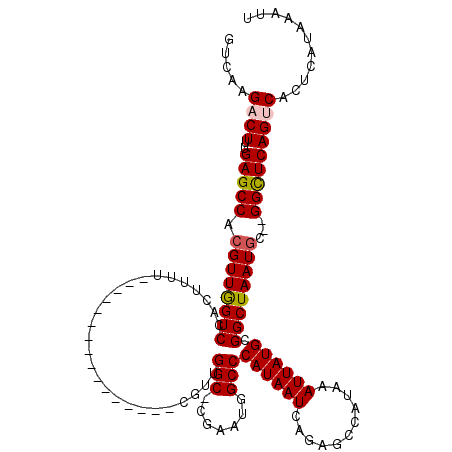
| Location | 10,122,054 – 10,122,155 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.96 |
| Mean single sequence MFE | -32.35 |
| Consensus MFE | -21.84 |
| Energy contribution | -22.12 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10122054 101 - 27905053 GUCAAGACUUUGAGCCACGUUGGUCCACUUUU----------------CAUUGGC-CGAAUGGCCCAUAAUCAGAGCCAUAAAUUAUGCGGCUAAUGC--GGUUCAGUCACUCAUAAAUU .....((((..(((((.((((((((.......----------------....(((-(....))))((((((...........)))))).)))))))).--)))))))))........... ( -28.90) >DroSec_CAF1 31320 102 - 1 GUCAAGACUUUGAGCCACGUUGGUCCACUUUU----------------CGUGGGCCCGAAUGGCCCAUAAUCAGAGCCAUAAAUUAUGCGGCUAAUGC--GGCUCAGUCACUCAUAAAUU .....((((..(((((.((((((((..((..(----------------..((((((.....))))))..)..)).((.(((...))))))))))))).--)))))))))........... ( -36.70) >DroSim_CAF1 27729 102 - 1 GUCAAGACUUUGAGCCAAGUUGGUCCACUUUU----------------CGUUGGCCCGAAUGGCCCAUAAUCAGAGCCAUAAAUUAUGCGGCUAAUGC--GGCUCAGUCACUCAUAAAUU .....((((..(((((...((((.(((.....----------------...))).)))).(((((((((((...........)))))).)))))....--)))))))))........... ( -30.00) >DroEre_CAF1 31064 101 - 1 GUCAAGACUCGGAGCCACGUUAGUCCACUUCU----------------CGUUGGC-CGAUUGGCCCAUAAUCAGAGCCAUAAAUUAUGCGGCUAAUGU--GGCUCAGGCACUCAUAAAUU ...........((((((((((((((((.....----------------...((((-((((((.....))))).).)))).......)).)))))))))--)))))............... ( -33.36) >DroYak_CAF1 31820 119 - 1 GUCAAGACUUUGAGCCACGUUGGUCAGCUUUUCGUUUUGUCGUUAUUUCGUUGGC-CGAAUGGCCCAUAAUCAGAGCCAUAAAUUAUGCGGCUAAUGUGCGGCUCAGGCAAUAAUAAAUU ........((((((((.(((((((((((....((......)).......))))))-))).(((((((((((...........)))))).)))))...)).))))))))............ ( -32.80) >consensus GUCAAGACUUUGAGCCACGUUGGUCCACUUUU________________CGUUGGC_CGAAUGGCCCAUAAUCAGAGCCAUAAAUUAUGCGGCUAAUGC__GGCUCAGUCACUCAUAAAUU .....((((..(((((.((((((((...........................(((.......)))((((((...........)))))).))))))))...)))))))))........... (-21.84 = -22.12 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:35 2006