| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,118,693 – 10,118,844 |
| Length | 151 |
| Max. P | 0.979152 |

| Location | 10,118,693 – 10,118,804 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.30 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -22.64 |
| Energy contribution | -23.87 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10118693 111 + 27905053 CAGCAAAAAAAAAAAAAUAAGAGCCAUCCA---UCUUUGUCCUUCGU--UUGAAAUAGACAUGUGAUAGGCUAAACAUGUACAUGGCAUGUAUAUUUCACUACCGACUGGCCCACU ....................(.((((....---.....(((....((--.(((((((.((((((.((..((.......))..)).)))))).)))))))..)).))))))).)... ( -21.20) >DroSec_CAF1 28026 98 + 1 CGGCCAA-----------AAGAGCCAUCCA---ACUUUGUCCUUCGCCAUUGAAAUAGACAUGCGAUAGGCCAAACA--UACAUGGCAUGU--AUUUCACUGGCGACUGGCCCACU .(((((.-----------.((((.......---.)))).....((((((.((((((..((((((.((.(........--..))).))))))--)))))).)))))).))))).... ( -31.90) >DroSim_CAF1 24465 101 + 1 CGGCCAA-----------AAGAGCCAUUUGUGUCCUGUGUCCUUCGCCAUUGAAAUAGACAUGCGAUAGGCCAAACA--UACAUGGCAUGU--AUUUCACUGGCGACUGGCCCACU .(((((.-----------.((..((....).)..)).......((((((.((((((..((((((.((.(........--..))).))))))--)))))).)))))).))))).... ( -32.20) >consensus CGGCCAA___________AAGAGCCAUCCA___ACUUUGUCCUUCGCCAUUGAAAUAGACAUGCGAUAGGCCAAACA__UACAUGGCAUGU__AUUUCACUGGCGACUGGCCCACU .(((((..............((....))...............((((((.((((((..((((((.((.(............))).))))))..)))))).)))))).))))).... (-22.64 = -23.87 + 1.23)


| Location | 10,118,693 – 10,118,804 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.30 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -24.46 |
| Energy contribution | -25.79 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10118693 111 - 27905053 AGUGGGCCAGUCGGUAGUGAAAUAUACAUGCCAUGUACAUGUUUAGCCUAUCACAUGUCUAUUUCAA--ACGAAGGACAAAGA---UGGAUGGCUCUUAUUUUUUUUUUUUUGCUG ...((((((..((((((.(...((.(((((.......))))).)).))))))...(((((..(((..--..))))))))....---.)..)))))).................... ( -24.70) >DroSec_CAF1 28026 98 - 1 AGUGGGCCAGUCGCCAGUGAAAU--ACAUGCCAUGUA--UGUUUGGCCUAUCGCAUGUCUAUUUCAAUGGCGAAGGACAAAGU---UGGAUGGCUCUU-----------UUGGCCG ....((((((((((((.((((((--((((((...(((--.(.....).))).))))))..)))))).)))))).......(((---(....))))...-----------)))))). ( -31.90) >DroSim_CAF1 24465 101 - 1 AGUGGGCCAGUCGCCAGUGAAAU--ACAUGCCAUGUA--UGUUUGGCCUAUCGCAUGUCUAUUUCAAUGGCGAAGGACACAGGACACAAAUGGCUCUU-----------UUGGCCG .(((..((..((((((.((((((--((((((...(((--.(.....).))).))))))..)))))).)))))).)).)))...........((((...-----------..)))). ( -32.80) >consensus AGUGGGCCAGUCGCCAGUGAAAU__ACAUGCCAUGUA__UGUUUGGCCUAUCGCAUGUCUAUUUCAAUGGCGAAGGACAAAGA___UGGAUGGCUCUU___________UUGGCCG ...((((((.((((((.((((((..((((((.(((.............))).))))))..)))))).)))))).................)))))).................... (-24.46 = -25.79 + 1.33)

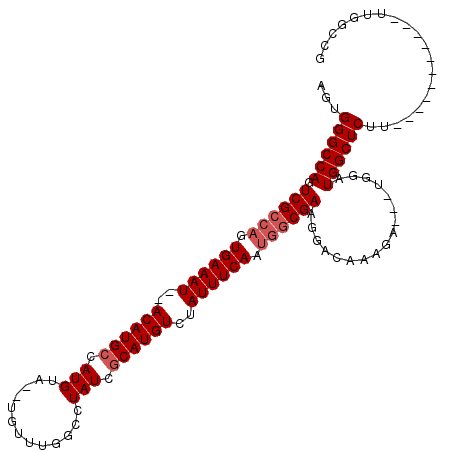
| Location | 10,118,730 – 10,118,844 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.86 |
| Mean single sequence MFE | -32.07 |
| Consensus MFE | -18.50 |
| Energy contribution | -20.93 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10118730 114 + 27905053 CCUUCGU--UUGAAAUAGACAUGUGAUAGGCUAAACAUGUACAUGGCAUGUA----UAUUUCACUACCGACUGGCCCACUUACGAGUACAUUUGAAUACAGGUGUAUCUUCGCCUGAUUG ..(((((--.(((((((.((((((.((..((.......))..)).)))))).----)))))))...((....)).......)))))............((((((......)))))).... ( -27.40) >DroSec_CAF1 28052 112 + 1 CCUUCGCCAUUGAAAUAGACAUGCGAUAGGCCAAACA--UACAUGGCAUGU------AUUUCACUGGCGACUGGCCCACUUACGAGUACAUUUGAAUACAGGUGUAUCUUCGCCUGAUUG ((.((((((.((((((..((((((.((.(........--..))).))))))------)))))).))))))..))........................((((((......)))))).... ( -33.40) >DroSim_CAF1 24494 112 + 1 CCUUCGCCAUUGAAAUAGACAUGCGAUAGGCCAAACA--UACAUGGCAUGU------AUUUCACUGGCGACUGGCCCACUUACGAGCACAUUUGAAUACAGGUGUAUCUUCGCCUGAUUG ((.((((((.((((((..((((((.((.(........--..))).))))))------)))))).))))))..))........................((((((......)))))).... ( -33.40) >DroEre_CAF1 27926 102 + 1 CCCUCGGCUGAG--------UGGCUGUAGGCCAAACU--UACAUGGCAUGCACGAGUACUUCACU--CGACUGGCACACUUA------CAUUUAAAUACAGGUGUAUCUUCGGCUGAUCG ...(((((((((--------((((((((.((((....--....)))).))).(((((.....)))--))...))).))).((------(((((......)))))))...))))))))... ( -34.10) >consensus CCUUCGCCAUUGAAAUAGACAUGCGAUAGGCCAAACA__UACAUGGCAUGU______AUUUCACUGGCGACUGGCCCACUUACGAGUACAUUUGAAUACAGGUGUAUCUUCGCCUGAUUG ((.((((((.((((((.........(((.((((..........)))).)))......)))))).))))))..))........................((((((......)))))).... (-18.50 = -20.93 + 2.44)

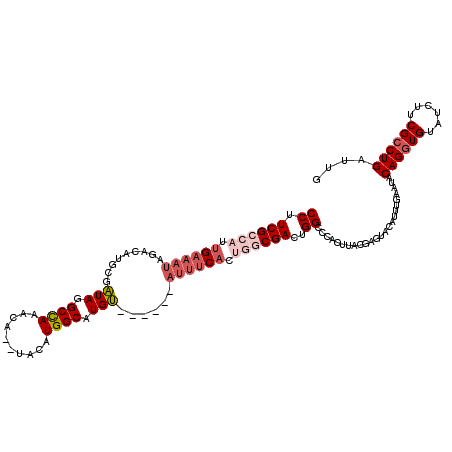
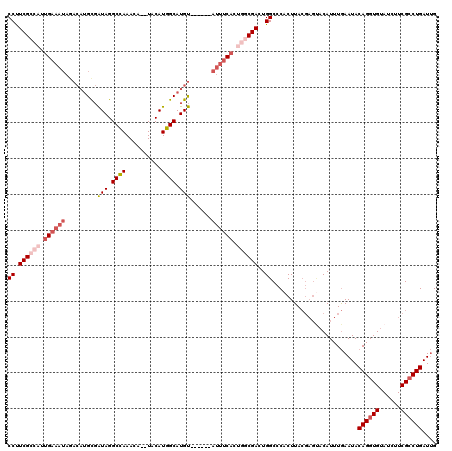
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:33 2006