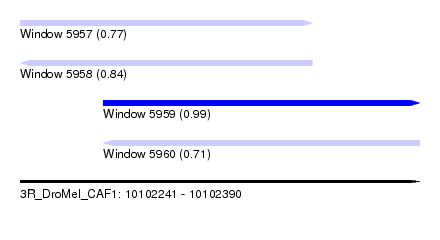
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,102,241 – 10,102,390 |
| Length | 149 |
| Max. P | 0.992835 |

| Location | 10,102,241 – 10,102,350 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 97.43 |
| Mean single sequence MFE | -39.82 |
| Consensus MFE | -36.20 |
| Energy contribution | -35.88 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10102241 109 + 27905053 AGUAAAGCGGCACUUAUAGGAAAGGUUCCACCUUGGGAGGAUCCUCCGGGGGAAACCAGGGCACAGGCCAAUCCAGUUGAUGUGGCCGGUGUCGCCACGAACAGUGCUU ....(((((((............((((((.(((..((((....))))))))).))))..(((((.((((((((.....))).))))).))))))))((.....)))))) ( -43.10) >DroSec_CAF1 11574 109 + 1 AGUAAAGCGGCACUUAUAGGAAAGGCUCCACCUUGCGACGAUCCUCCGGGGGAAACCAGGGCACAGGCCAAUCCAGUUGAUGUGGCCGGUGUCGCCACGAAUAGUGCUU ........((((((....(....(((..((((..((.((.(((..(.((((....))..(((....)))...)).)..))))).)).))))..))).)....)))))). ( -37.20) >DroSim_CAF1 7772 109 + 1 AGUAAAGCGGCACUUAUAGGAAAGGCUCCACCUUGCGACGAUCCUCCGGGGGAAACCAGGGCACAGGCCAAUCCAGUUGAUGUGGCCGGUGUCGCCACGAAUAGUGCUU ........((((((....(....(((..((((..((.((.(((..(.((((....))..(((....)))...)).)..))))).)).))))..))).)....)))))). ( -37.20) >DroEre_CAF1 11639 109 + 1 AGUAAAGCGGCACUUAUAGGAAAGGCUCCACCUUGCGAGGAUCCUCCUGGGGAAACCAGGGCACAGGCCAAUCCAGUUGAUGUGGCCGGUGUCGCCACGAAUAGUGCUU ........((((((....(....(((..((((.((((((....)))((((.....)))).)))..((((((((.....))).)))))))))..))).)....)))))). ( -40.80) >DroYak_CAF1 11650 109 + 1 AGUAAAGCGGCACUUAUAGGAAAGGCUCCACCUUGCGAGGAUCCUCCUGGGGAAACCAGGGCACAGGCCAGUCCAGUUGAUGUGGCCGGUGUCGCCACGAAUAGUGCUU ........((((((....(....(((..((((.((((((....)))((((.....)))).)))..((((((((.....))).)))))))))..))).)....)))))). ( -40.80) >consensus AGUAAAGCGGCACUUAUAGGAAAGGCUCCACCUUGCGAGGAUCCUCCGGGGGAAACCAGGGCACAGGCCAAUCCAGUUGAUGUGGCCGGUGUCGCCACGAAUAGUGCUU ........((((((....(....(((..((((.............((..((....))..))....((((((((.....))).)))))))))..))).)....)))))). (-36.20 = -35.88 + -0.32)

| Location | 10,102,241 – 10,102,350 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 97.43 |
| Mean single sequence MFE | -39.46 |
| Consensus MFE | -33.10 |
| Energy contribution | -33.54 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10102241 109 - 27905053 AAGCACUGUUCGUGGCGACACCGGCCACAUCAACUGGAUUGGCCUGUGCCCUGGUUUCCCCCGGAGGAUCCUCCCAAGGUGGAACCUUUCCUAUAAGUGCCGCUUUACU ..(((((......((.(.(((.(((((.(((.....)))))))).)))))).((((((((..((((....))))...)).)))))).........)))))......... ( -38.70) >DroSec_CAF1 11574 109 - 1 AAGCACUAUUCGUGGCGACACCGGCCACAUCAACUGGAUUGGCCUGUGCCCUGGUUUCCCCCGGAGGAUCGUCGCAAGGUGGAGCCUUUCCUAUAAGUGCCGCUUUACU ..(((((....((((((((((.(((((.(((.....)))))))).)))((((((......)))).)).)))))))((((.....)))).......)))))......... ( -39.70) >DroSim_CAF1 7772 109 - 1 AAGCACUAUUCGUGGCGACACCGGCCACAUCAACUGGAUUGGCCUGUGCCCUGGUUUCCCCCGGAGGAUCGUCGCAAGGUGGAGCCUUUCCUAUAAGUGCCGCUUUACU ..(((((....((((((((((.(((((.(((.....)))))))).)))((((((......)))).)).)))))))((((.....)))).......)))))......... ( -39.70) >DroEre_CAF1 11639 109 - 1 AAGCACUAUUCGUGGCGACACCGGCCACAUCAACUGGAUUGGCCUGUGCCCUGGUUUCCCCAGGAGGAUCCUCGCAAGGUGGAGCCUUUCCUAUAAGUGCCGCUUUACU ..(((((((..(.(((..(((((((((.(((.....))))))))..(((.((((.....))))(((....)))))).))))..)))...)..)).)))))......... ( -40.50) >DroYak_CAF1 11650 109 - 1 AAGCACUAUUCGUGGCGACACCGGCCACAUCAACUGGACUGGCCUGUGCCCUGGUUUCCCCAGGAGGAUCCUCGCAAGGUGGAGCCUUUCCUAUAAGUGCCGCUUUACU ..(((((((..(.(((..(((((((((..((....))..)))))..(((.((((.....))))(((....)))))).))))..)))...)..)).)))))......... ( -38.70) >consensus AAGCACUAUUCGUGGCGACACCGGCCACAUCAACUGGAUUGGCCUGUGCCCUGGUUUCCCCCGGAGGAUCCUCGCAAGGUGGAGCCUUUCCUAUAAGUGCCGCUUUACU ..(((((((..(.(((..(((((((((.(((.....))))))))..(((...((((..(....)..))))...))).))))..)))...)..)).)))))......... (-33.10 = -33.54 + 0.44)


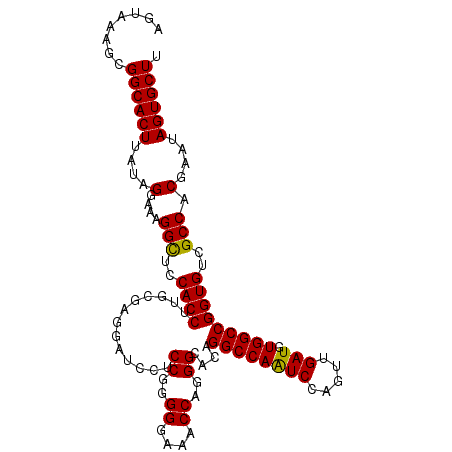
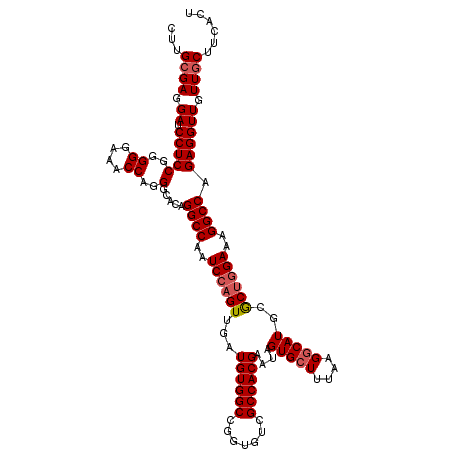
| Location | 10,102,272 – 10,102,390 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 95.25 |
| Mean single sequence MFE | -48.14 |
| Consensus MFE | -41.80 |
| Energy contribution | -42.36 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10102272 118 + 27905053 CUUGGGAGGAUCCUCCGGGGGAAACCAGGGCACAGGCCAAUCCAGUUGAUGUGGCCGGUGUCGCCACGAACAGUGCUUUGAGACAUGCGCCUGAAAGGCCAGAGGUUGUUGCUUCACU (((.((((....)))).))).....((((((((.((((((((.....))).)))))((.....)).......)))))))).....((.(((.....)))))(((((....)))))... ( -43.10) >DroSec_CAF1 11605 118 + 1 CUUGCGACGAUCCUCCGGGGGAAACCAGGGCACAGGCCAAUCCAGUUGAUGUGGCCGGUGUCGCCACGAAUAGUGCUUUAAGGCAUGCGCUGGAAAGGCCAGAGGUUGUUGCUUCACU ...(((((((.((((.((((....))..(((((.((((((((.....))).))))).))))).(((((....(((((....))))).)).))).....)).)))))))))))...... ( -49.80) >DroSim_CAF1 7803 118 + 1 CUUGCGACGAUCCUCCGGGGGAAACCAGGGCACAGGCCAAUCCAGUUGAUGUGGCCGGUGUCGCCACGAAUAGUGCUUUAAGGCAUGCGCUGGAAAGGCCAGAGGUUGUUGCUUCACU ...(((((((.((((.((((....))..(((((.((((((((.....))).))))).))))).(((((....(((((....))))).)).))).....)).)))))))))))...... ( -49.80) >DroEre_CAF1 11670 118 + 1 CUUGCGAGGAUCCUCCUGGGGAAACCAGGGCACAGGCCAAUCCAGUUGAUGUGGCCGGUGUCGCCACGAAUAGUGCUUUAAGGCAUGCACUGGAAAGGCCAGAGGUUGUUGCUUCACA ...((((.(((((((((((.....))))).....((((..((((((...((((((.......))))))....(((((....)))))..))))))..)))))).)))).))))...... ( -47.60) >DroYak_CAF1 11681 118 + 1 CUUGCGAGGAUCCUCCUGGGGAAACCAGGGCACAGGCCAGUCCAGUUGAUGUGGCCGGUGUCGCCACGAAUAGUGCUUUAAGGCAUGAACUGGAAAGGCCAGAGGUUGUUGCUUCACA ...((((.(((((((((((.....))))).....((((..(((((((..((((((.......))))))....(((((....))))).)))))))..)))))).)))).))))...... ( -50.40) >consensus CUUGCGAGGAUCCUCCGGGGGAAACCAGGGCACAGGCCAAUCCAGUUGAUGUGGCCGGUGUCGCCACGAAUAGUGCUUUAAGGCAUGCGCUGGAAAGGCCAGAGGUUGUUGCUUCACU ...((((.((.(((((..((....))..).....((((..((((((...((((((.......))))))....(((((....)))))..))))))..)))).)))))).))))...... (-41.80 = -42.36 + 0.56)


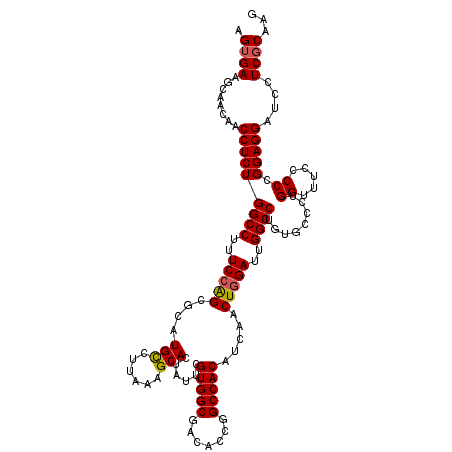
| Location | 10,102,272 – 10,102,390 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 95.25 |
| Mean single sequence MFE | -40.32 |
| Consensus MFE | -33.86 |
| Energy contribution | -33.94 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10102272 118 - 27905053 AGUGAAGCAACAACCUCUGGCCUUUCAGGCGCAUGUCUCAAAGCACUGUUCGUGGCGACACCGGCCACAUCAACUGGAUUGGCCUGUGCCCUGGUUUCCCCCGGAGGAUCCUCCCAAG .((......))..(((((((....(((((.((((.................(((....))).(((((.(((.....)))))))).)))))))))......)))))))........... ( -37.70) >DroSec_CAF1 11605 118 - 1 AGUGAAGCAACAACCUCUGGCCUUUCCAGCGCAUGCCUUAAAGCACUAUUCGUGGCGACACCGGCCACAUCAACUGGAUUGGCCUGUGCCCUGGUUUCCCCCGGAGGAUCGUCGCAAG ......((.((..(((((((.....((((.((((((......)).......(((....))).(((((.(((.....)))))))).)))).))))......)))))))...)).))... ( -40.00) >DroSim_CAF1 7803 118 - 1 AGUGAAGCAACAACCUCUGGCCUUUCCAGCGCAUGCCUUAAAGCACUAUUCGUGGCGACACCGGCCACAUCAACUGGAUUGGCCUGUGCCCUGGUUUCCCCCGGAGGAUCGUCGCAAG ......((.((..(((((((.....((((.((((((......)).......(((....))).(((((.(((.....)))))))).)))).))))......)))))))...)).))... ( -40.00) >DroEre_CAF1 11670 118 - 1 UGUGAAGCAACAACCUCUGGCCUUUCCAGUGCAUGCCUUAAAGCACUAUUCGUGGCGACACCGGCCACAUCAACUGGAUUGGCCUGUGCCCUGGUUUCCCCAGGAGGAUCCUCGCAAG (((((.(((.((......((((..((((((...(((......)))......(((((.......)))))....))))))..)))))))))(((((.....))))).......))))).. ( -42.20) >DroYak_CAF1 11681 118 - 1 UGUGAAGCAACAACCUCUGGCCUUUCCAGUUCAUGCCUUAAAGCACUAUUCGUGGCGACACCGGCCACAUCAACUGGACUGGCCUGUGCCCUGGUUUCCCCAGGAGGAUCCUCGCAAG (((((.(((.((......((((..(((((((..(((......)))......(((((.......)))))...)))))))..)))))))))(((((.....))))).......))))).. ( -41.70) >consensus AGUGAAGCAACAACCUCUGGCCUUUCCAGCGCAUGCCUUAAAGCACUAUUCGUGGCGACACCGGCCACAUCAACUGGAUUGGCCUGUGCCCUGGUUUCCCCCGGAGGAUCCUCGCAAG .((((........(((((((((..(((((....(((......)))......(((((.......))))).....)))))..))))........((.....)).)))))....))))... (-33.86 = -33.94 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:20 2006