
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,100,018 – 10,100,114 |
| Length | 96 |
| Max. P | 0.936281 |

| Location | 10,100,018 – 10,100,114 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 81.32 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -23.84 |
| Energy contribution | -25.42 |
| Covariance contribution | 1.57 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
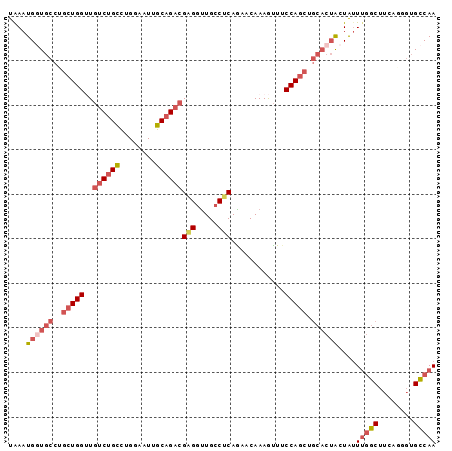
>3R_DroMel_CAF1 10100018 96 + 27905053 UAAAUGUUGCCUGCUGGUUGUCUGCCUGGAAUUGCAGACGAGGUUGCCUCAGGACAAAGUUUCCAGCUGCACUACUAUUUGGCUUCAAGGUGCCAA .......(((..(((((..((((((........))))))((((...))))............))))).))).......(((((........))))) ( -30.40) >DroVir_CAF1 13634 81 + 1 CAAAUGAUCCGCUC---UUAUCCGUUUGGAAUUGCAGCCGCGGCUGGCGCAGCACAAAGCU------------GCUGUUUACCUUCGCGGUGGCAA .......((((..(---......)..)))).((((.(((((((..((.((((((......)------------)))))...)).))))))).)))) ( -28.30) >DroSec_CAF1 9317 96 + 1 UGAAUGGUGCCUGCUGGUUGUCUGCCUGGAAUUGCAGACGAGGUUGCCUCAAAACAAAGUUUCCAGCUGCACUACUAUUUGGCUUCAGGGUGCCAA ....((((((..(((((..((((((........))))))((((...))))............))))).))))))....(((((........))))) ( -35.80) >DroSim_CAF1 5506 96 + 1 UGAAUGGUGCCUGCUGGUUGUCUGCCUGGAAUUGCAGACGAGGUUGCCUCAAAACAAAGUUUCCAGCUGCACUACUAUUUGGCUUCAGGGUGCCAA ....((((((..(((((..((((((........))))))((((...))))............))))).))))))....(((((........))))) ( -35.80) >DroEre_CAF1 9360 96 + 1 UAAAUGGUGCCGGCUGGGUGUCUGCCUGGAAUUGCAGACGAGGUUGCCUCAGUACAAAAUUUCCAGCUGCACUGCUAUUUGGCUUCAGGUUGCCAA .....(((((.(((((((.((((((........))))))((((...))))...........)))))))))))).....(((((........))))) ( -37.00) >DroYak_CAF1 9369 96 + 1 UAAAUGGUGCCUGCUGGGUGUCUGCCUGGAAUUGCAGACGUGGUUGCCUCAGAACAAAAUUUCCAGCUGCACUGCUACUGGGCUUCAGGGUGCCAA ....(((..((((((.(((((((((........))))))(..((((.....(((......)))))))..)......))).)))....)))..))). ( -33.00) >consensus UAAAUGGUGCCUGCUGGUUGUCUGCCUGGAAUUGCAGACGAGGUUGCCUCAGAACAAAGUUUCCAGCUGCACUACUAUUUGGCUUCAGGGUGCCAA ....((((((..(((((..((((((........))))))(((.....)))............))))).))))))....(((((........))))) (-23.84 = -25.42 + 1.57)


| Location | 10,100,018 – 10,100,114 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 81.32 |
| Mean single sequence MFE | -31.85 |
| Consensus MFE | -24.66 |
| Energy contribution | -25.70 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
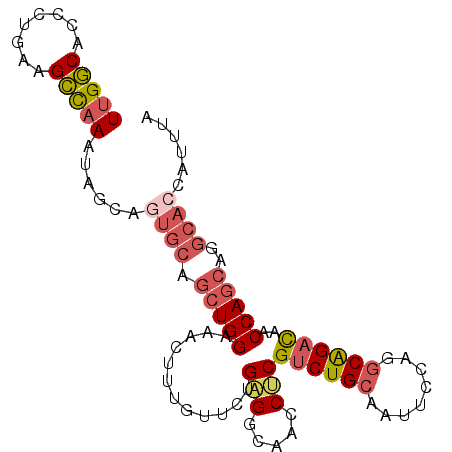
>3R_DroMel_CAF1 10100018 96 - 27905053 UUGGCACCUUGAAGCCAAAUAGUAGUGCAGCUGGAAACUUUGUCCUGAGGCAACCUCGUCUGCAAUUCCAGGCAGACAACCAGCAGGCAACAUUUA (((((........))))).......(((.(((((............((((...))))((((((........))))))..)))))..)))....... ( -32.00) >DroVir_CAF1 13634 81 - 1 UUGCCACCGCGAAGGUAAACAGC------------AGCUUUGUGCUGCGCCAGCCGCGGCUGCAAUUCCAAACGGAUAA---GAGCGGAUCAUUUG ......((((...........((------------((((.((.((((...)))))).))))))...(((....)))...---..))))........ ( -26.00) >DroSec_CAF1 9317 96 - 1 UUGGCACCCUGAAGCCAAAUAGUAGUGCAGCUGGAAACUUUGUUUUGAGGCAACCUCGUCUGCAAUUCCAGGCAGACAACCAGCAGGCACCAUUCA (((((........)))))......((((.(((((............((((...))))((((((........))))))..)))))..))))...... ( -34.70) >DroSim_CAF1 5506 96 - 1 UUGGCACCCUGAAGCCAAAUAGUAGUGCAGCUGGAAACUUUGUUUUGAGGCAACCUCGUCUGCAAUUCCAGGCAGACAACCAGCAGGCACCAUUCA (((((........)))))......((((.(((((............((((...))))((((((........))))))..)))))..))))...... ( -34.70) >DroEre_CAF1 9360 96 - 1 UUGGCAACCUGAAGCCAAAUAGCAGUGCAGCUGGAAAUUUUGUACUGAGGCAACCUCGUCUGCAAUUCCAGGCAGACACCCAGCCGGCACCAUUUA (((((........)))))......((((.(((((............((((...))))((((((........))))))..)))))..))))...... ( -34.50) >DroYak_CAF1 9369 96 - 1 UUGGCACCCUGAAGCCCAGUAGCAGUGCAGCUGGAAAUUUUGUUCUGAGGCAACCACGUCUGCAAUUCCAGGCAGACACCCAGCAGGCACCAUUUA ..(((........)))........((((.(((((...........((.(....))).((((((........))))))..)))))..))))...... ( -29.20) >consensus UUGGCACCCUGAAGCCAAAUAGCAGUGCAGCUGGAAACUUUGUUCUGAGGCAACCUCGUCUGCAAUUCCAGGCAGACAACCAGCAGGCACCAUUUA (((((........)))))......((((.(((((............(((.....)))((((((........))))))..)))))..))))...... (-24.66 = -25.70 + 1.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:16 2006