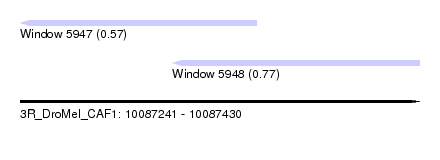
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,087,241 – 10,087,430 |
| Length | 189 |
| Max. P | 0.772464 |

| Location | 10,087,241 – 10,087,353 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.50 |
| Mean single sequence MFE | -36.73 |
| Consensus MFE | -21.86 |
| Energy contribution | -22.53 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10087241 112 - 27905053 UUACGACAAGACCAAGAAGGCUGCCUAUAAGGCGGCUUGUGCGGCAGUGGAGGCCAAGCAAAAGGCUCAAAGGAUGCAAAGGGCGA--------UUAGCUCUGCAUCCUCUUCAAGCUAG ............((...((((((((.....)))))))).)).(((..((((((((........)))....((((((((..((((..--------...))))))))))))))))).))).. ( -41.20) >DroVir_CAF1 198950 119 - 1 CUUCGAGAAGACCAAAAAGGCCGCCUACAAAGCAGCCUGUUCGGCGGUCGAGGCCAAGCAGAAGGCGCAAAGGAGUCGACGGCGAACCGGUGGACCCGCACAGGGUUCUCUUUAA-UUAU ....((((..(((....((((.((.......)).))))(((((.(.(((((..((..((.......))...))..))))).)))))).)))((((((.....))))))))))...-.... ( -40.90) >DroSec_CAF1 145970 112 - 1 UUACGAAAAGACCAAGCAGGCUGCCUACAAGGCGGCUUGUGCGGCAGUGGAGGCCAAGAAAAAGGCUCAAAUGAUGCAAACGUAGA--------UCAGCUCUGCAUCCUCUUCAAGUUAG .........(.((..((((((((((.....))))))))))..))).(.(((((((........)))......((((((...((...--------...))..)))))))))).)....... ( -35.20) >DroSim_CAF1 147876 112 - 1 UUACGAAAAGACCAAGCAGGCUGCCUACAAGGCGGCUUGUGCGGCAGUGGAGGCCAAGAAAAAGGCUCAAAUGAUGCAAACGUAGA--------UCAGCUCUGCAUCCUCUUCAAGUUAG .........(.((..((((((((((.....))))))))))..))).(.(((((((........)))......((((((...((...--------...))..)))))))))).)....... ( -35.20) >DroEre_CAF1 158291 111 - 1 UUACGACAAGACCAAGAAGGCUGCCUACAAGGCGGCUUGUGCGGCAGUGGAGGCCAAGCAAAAGGCUCAAAGGAUGCAAAGGGAAA--------UCAGCUCCGCCUCC-CUUCAAGCUAA ...............((((((((((((((((....)))))).))))))((((((..(((.....)))....(((.((.........--------...)))))))))))-))))....... ( -37.80) >DroYak_CAF1 153226 111 - 1 UUACGACAAGACCAAGAAAGCUGCCUACAAGGCGGCUUGUGCGGCAGUGGAGGCCAAGCAAAAGGCUCAAAGGAUGCAAAGGGAAA--------UCAGCGAGGCAGAG-CUUGAAGCUAA ...(..((...((....((((((((.....))))))))....))...))..)....(((...((((((....(((..........)--------)).((...)).)))-)))...))).. ( -30.10) >consensus UUACGACAAGACCAAGAAGGCUGCCUACAAGGCGGCUUGUGCGGCAGUGGAGGCCAAGCAAAAGGCUCAAAGGAUGCAAAGGGAAA________UCAGCUCUGCAUCCUCUUCAAGCUAG .........(.((....((((((((.....))))))))....)))..((((((((........)))......(((((.........................)))))..)))))...... (-21.86 = -22.53 + 0.67)


| Location | 10,087,313 – 10,087,430 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.68 |
| Mean single sequence MFE | -44.45 |
| Consensus MFE | -28.09 |
| Energy contribution | -28.12 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10087313 117 - 27905053 GAGAAGGGGCAGCUGUUGGAGGCGGCCAAUCGGAGAGUGGAGAGCAUCUCCAGACAAGUGGUCGCUGCCAGGCAGGAUUACGACAAGACCAAGAAGGCUGCCUAUAAGGCGGCUUGU ........(((((((((..((((((((....(((((((.....)).)))))......((((((.((((...))))))))))..............))))))))....)))))).))) ( -45.80) >DroVir_CAF1 199029 117 - 1 GAGAAGACGCAGCUGCUGGAAGCGGCCAAGAAUCGUGUGGAGAGCUUAGCCCGCCAGCUCGCCUUGGCCAAGCAGGACUUCGAGAAGACCAAAAAGGCCGCCUACAAAGCAGCCUGU ........(((((((((....((((((................((((.(((.((......))...))).)))).((.(((....))).)).....))))))......))))).)))) ( -38.90) >DroSec_CAF1 146042 117 - 1 GAGAAGGGUCAGCUGUUGGAGGCGGCCAAUCGGAGAGUGGAGGGCAUCUCCAGGCAAGUGGGCGCUGCCAGGCAGGAUUACGAAAAGACCAAGCAGGCUGCCUACAAGGCGGCUUGU ......((((.(((..(((.((((.(((.(((((((((.....)).))))).))....))).)))).)))))).(.....).....))))..((((((((((.....)))))))))) ( -47.90) >DroSim_CAF1 147948 117 - 1 GAGAAGGGUCAGCUGUUGGAGGCGGCCAAUCGGAGAGUGGAGGGCAUCUCCAGGCAAGUGGGCGCUGCCAGGCAGGAUUACGAAAAGACCAAGCAGGCUGCCUACAAGGCGGCUUGU ......((((.(((..(((.((((.(((.(((((((((.....)).))))).))....))).)))).)))))).(.....).....))))..((((((((((.....)))))))))) ( -47.90) >DroEre_CAF1 158362 117 - 1 GAAAAGGGGCAUCUGCUGGAGGCUGCCAAUCGGAGAAUGGAGUGCGUCUACAGGCAGGUCCUCGCUGCCAGGCAGGAUUACGACAAGACCAAGAAGGCUGCCUACAAGGCGGCUUGU .....((((.((((((((((.((..(((.((...)).)))...)).)))...)))))))))))((((((((((((.....(.....).((.....))))))))....)))))).... ( -43.80) >DroMoj_CAF1 177781 117 - 1 GAGAAGACGCAGCUGCUGGAGGCGGCCAAGAAUCGAGUGGACAGCCUAUUGCGUCAGCUGAACUUGGCCAAGCAGGACUACGAUAAGACCAAGAAGGCGGCCUACAAAGCAGCCUGC ........(((((((((..(((((((((((..((.(((.(((.((.....))))).))))).))))))).....((.((......)).)).........))))....))))).)))) ( -42.40) >consensus GAGAAGGGGCAGCUGCUGGAGGCGGCCAAUCGGAGAGUGGAGAGCAUCUCCAGGCAAGUGGGCGCUGCCAGGCAGGAUUACGAAAAGACCAAGAAGGCUGCCUACAAGGCGGCUUGU ........(((((((((..((((((((.......(..(((...((..................))..)))..).((.((......)).)).....))))))))....)))))).))) (-28.09 = -28.12 + 0.03)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:08 2006