
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,084,149 – 10,084,291 |
| Length | 142 |
| Max. P | 0.731041 |

| Location | 10,084,149 – 10,084,258 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 82.31 |
| Mean single sequence MFE | -36.42 |
| Consensus MFE | -22.99 |
| Energy contribution | -25.89 |
| Covariance contribution | 2.89 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.731041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
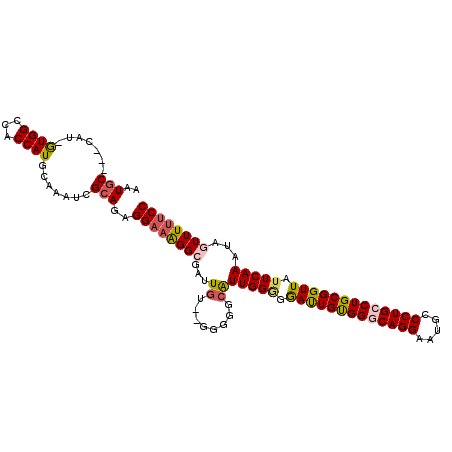
>3R_DroMel_CAF1 10084149 109 - 27905053 GA-AGGCGAUUGU--GGGGGAAUUGGGGGAUUGUGGGCAGGAAUGCCCUGCCUGCGGUUAUUCAAAUAGUUUUUCCAUCAAAUUAUCAACCGAAACUGUU-GAGGUUGCCUUG .(-((((((((((--((..((((((.(((((((..((((((.....))))))..)))))..))...))))))..)))).......(((((.......)))-))))))))))). ( -44.80) >DroPse_CAF1 144176 89 - 1 ----GGUGGUU---------CCUUGGA-------GGGGAGAGAUG----GCCUGCGGUUAUUCAAAUAGUUUUUCCAUCAAAUCAUCAGCAAAAACUGUUUGGGGUUGUCUUG ----.....((---------(((....-------)))))((((..----((((.........((((((((((((.(............).))))))))))))))))..)))). ( -23.00) >DroSec_CAF1 142928 110 - 1 GAAAGGCGAUUGU--GGGGGCAUUGGGGGAUUGUGGGCAGGAAUGCCCUGCCUGCGGUUAUUCAAAUAGUUUUUCCAUCAAAUUAUCAACCGAAACUGUU-GAGGUUGCCUUG ..(((((((((((--((..(.((((.(((((((..((((((.....))))))..)))))..))...)))).)..)))).......(((((.......)))-))))))))))). ( -40.80) >DroSim_CAF1 144847 110 - 1 GAAAGGCGAUUGU--GGGGGCAUUGGGGGAUUGUGGGCAGGAAUGCCCUGCCUGCGGUUAUUCAAAUAGUUUUUCCAUCAAAUUAUCAACCGAAACUGUU-GAGGUUGCCUUG ..(((((((((((--((..(.((((.(((((((..((((((.....))))))..)))))..))...)))).)..)))).......(((((.......)))-))))))))))). ( -40.80) >DroEre_CAF1 155198 108 - 1 GCAGGG--AAUGA--AUGGGCGUUGGAGGACUGCGGGCAGGAAUGCCCUGGCUGCGGUUAUUCAAAUAGUUUUUCCAUCAAAUUAUCAACCGAAACUGUU-GAGGCUGCCUUG ((((((--((.((--((.....(((((.((((((((.((((.....)))).)))))))).)))))...)))))))).........(((((.......)))-))..)))).... ( -38.10) >DroYak_CAF1 150166 110 - 1 GAAGGG--UAUGGGCAUUGGCGUUGGCGAAUUGUGGGCAGGAAUGCCCUGGCUGCGGUUAUUCAAAUAGUUUUUCCAUCAAAUUAUCAACCGAAACUGUU-GAGGUUGCCUUG .(((((--.(((((....(((.((((..(((((..(.((((.....)))).)..)))))..))))...)))..)))))).......(((((.........-..))))))))). ( -31.00) >consensus GAAAGGCGAUUGU__GGGGGCAUUGGGGGAUUGUGGGCAGGAAUGCCCUGCCUGCGGUUAUUCAAAUAGUUUUUCCAUCAAAUUAUCAACCGAAACUGUU_GAGGUUGCCUUG ..(((((((((...........(((((.(((((((((((((.....))))))))))))).)))))((((((((..................))))))))....))))))))). (-22.99 = -25.89 + 2.89)



| Location | 10,084,185 – 10,084,291 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 85.95 |
| Mean single sequence MFE | -36.80 |
| Consensus MFE | -26.66 |
| Energy contribution | -27.22 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10084185 106 - 27905053 AAUGC---CACCAUGGACACCAUGCAAAUCGCAAAGGA-AGGCGAUUGU--GGGGGAAUUGGGGGAUUGUGGGCAGGAAUGCCCUGCCUGCGGUUAUUCAAAUAGUUUUUCC ....(---((((((((...)))))..((((((......-..))))))))--))((((((((((.(((((..((((((.....))))))..))))).)))))......))))) ( -39.40) >DroSec_CAF1 142964 106 - 1 AAUGC---CAU-GUGGCCACCAUGCAAAUCGCAGAGGAAAGGCGAUUGU--GGGGGCAUUGGGGGAUUGUGGGCAGGAAUGCCCUGCCUGCGGUUAUUCAAAUAGUUUUUCC ..(.(---((.-(((.((.((((...((((((.........))))))))--)))).)))))).)(((((..((((((.....))))))..)))))................. ( -39.70) >DroSim_CAF1 144883 106 - 1 AAUGC---CAU-GUGGCCACCAUGCAAAUCGCAGAGGAAAGGCGAUUGU--GGGGGCAUUGGGGGAUUGUGGGCAGGAAUGCCCUGCCUGCGGUUAUUCAAAUAGUUUUUCC ..(.(---((.-(((.((.((((...((((((.........))))))))--)))).)))))).)(((((..((((((.....))))))..)))))................. ( -39.70) >DroEre_CAF1 155234 107 - 1 AAUGCCACCAU-GUGGCCACCAUGAAAAUCGCAGAGGCAGGG--AAUGA--AUGGGCGUUGGAGGACUGCGGGCAGGAAUGCCCUGGCUGCGGUUAUUCAAAUAGUUUUUCC ...(((((...-)))))..((((.....((((....)).)).--.....--))))((.(((((.((((((((.((((.....)))).)))))))).)))))...))...... ( -36.20) >DroYak_CAF1 150202 106 - 1 AAUGC---CAU-GUGGCUACCAUGAAAAGCGGAGAGGAAGGG--UAUGGGCAUUGGCGUUGGCGAAUUGUGGGCAGGAAUGCCCUGGCUGCGGUUAUUCAAAUAGUUUUUCC (((((---((.-...((..(((((.....(......).....--)))))))..)))))))((..(((((..(.((((.....)))).)..)))))..))............. ( -29.00) >consensus AAUGC___CAU_GUGGCCACCAUGCAAAUCGCAGAGGAAAGGCGAUUGU__GGGGGCAUUGGGGGAUUGUGGGCAGGAAUGCCCUGCCUGCGGUUAUUCAAAUAGUUUUUCC ..(((.......((((...)))).......)))..((((((((...((........))(((((.(((((((((((((.....))))))))))))).)))))...)))))))) (-26.66 = -27.22 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:59 2006