| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 293,061 – 293,163 |
| Length | 102 |
| Max. P | 0.995622 |

| Location | 293,061 – 293,163 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 72.11 |
| Mean single sequence MFE | -45.68 |
| Consensus MFE | -20.08 |
| Energy contribution | -19.87 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.44 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
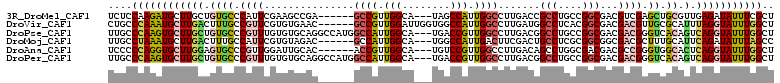
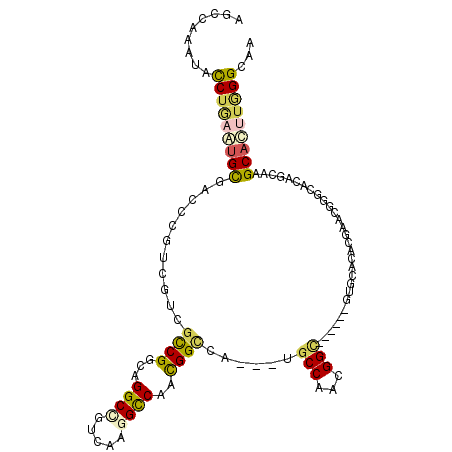
>3R_DroMel_CAF1 293061 102 + 27905053 UCUCCAAGAUGCUUGCUGUGCCCAUUCGAAGCCGA------GCCGUUGGCA---UAGCCAUUGGCCUUGACCGCCUGCCGGCGACGUCGAGCUGCGUUGAGAUAUUUCGCU ((((...(((((..((((((((..((((....)))------).....))))---))))....(((.(((((((((....))))..)))))))))))))))))......... ( -35.40) >DroVir_CAF1 57407 105 + 1 CUGCCCAAAUGCUUGACUUUGCCGUUCGUGUGAAC------GCCGUUGGAUUGGUGGCCAUUGGCCUUGAUGGCCUCACGGCGACGACGUUGCGCAUUUAGGUAUUUGGCU ..(((.((((((((((...(((....((((((..(------((((......)))))..))..((((.....)))).))))(((((...)))))))).))))))))))))). ( -39.80) >DroPse_CAF1 32399 108 + 1 UUGCCCAAGUGCUUGCUGUGCCCGUUUGUGUGCAGGCCAUGGCCAUUGGCA---UGACCGUUGGCCUUGACGGCCUGCCGGCGACGACGGGUCACAGUCAGGUAUUUGGCU ..(((.(((((((((((((((((((((((..(((((((..(((((.(((..---...))).))))).....)))))))..)))..)))))).))))).)))))))))))). ( -60.90) >DroMoj_CAF1 42578 102 + 1 UUGCCUAAAUGCUUGACUUUGCCAUUCGUGUAGAC------GCCAUUGGCA---UGGCCAUUGACUUCGACUGCCUCGCGGCGGCGACGCUUUGCAUUCAGAUAUUUAGCC ....(((((((.((((...(((((...(((....)------))...)))))---.(((..(((....)))..)))..((((((....))))..))..)))).))))))).. ( -29.60) >DroAna_CAF1 29119 102 + 1 UCCCCCAGGUGCUUGGAGUGCCCGUUGGAUUGCAC------ACCGUUGGCA---UGUCCGUUGGCCUUGACAGCCUGGCGACGACGCCGGGUGGCACUCAGGUAUUUGGCU ....(((((((((((.((((((((.((((((((.(------(....)))))---.))))).)).........((((((((....))))))))))))))))))))))))).. ( -47.50) >DroPer_CAF1 32838 108 + 1 UUGCCCAAGUGCUUGCUGUGCCCGUUUGUGUGCAGGCCAUGGCCAUUGGCA---UGACCGUUGGCCUUGACGGCCUGCCGGCGACGACGGGUCACAGUCAGGUAUUUGGCU ..(((.(((((((((((((((((((((((..(((((((..(((((.(((..---...))).))))).....)))))))..)))..)))))).))))).)))))))))))). ( -60.90) >consensus UUGCCCAAAUGCUUGCUGUGCCCGUUCGUGUGCAC______GCCAUUGGCA___UGGCCAUUGGCCUUGACGGCCUGCCGGCGACGACGGGUCGCAUUCAGGUAUUUGGCU ....((((((((((((.((((.((((...............((((.(((........))).)))).......(((....)))...)))).)).)).).))))))))))).. (-20.08 = -19.87 + -0.22)
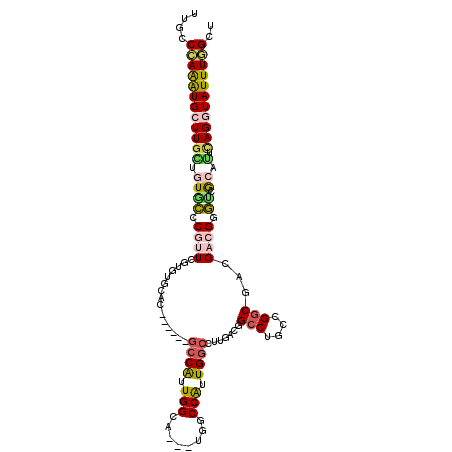
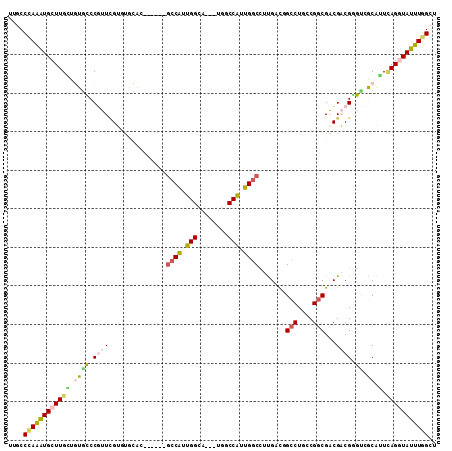
| Location | 293,061 – 293,163 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 72.11 |
| Mean single sequence MFE | -40.58 |
| Consensus MFE | -18.80 |
| Energy contribution | -18.92 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.46 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 293061 102 - 27905053 AGCGAAAUAUCUCAACGCAGCUCGACGUCGCCGGCAGGCGGUCAAGGCCAAUGGCUA---UGCCAACGGC------UCGGCUUCGAAUGGGCACAGCAAGCAUCUUGGAGA .........((((......(((((.((..((((((.(..((.((.((((...)))).---))))..).))------.))))..))..)))))....((((...)))))))) ( -31.70) >DroVir_CAF1 57407 105 - 1 AGCCAAAUACCUAAAUGCGCAACGUCGUCGCCGUGAGGCCAUCAAGGCCAAUGGCCACCAAUCCAACGGC------GUUCACACGAACGGCAAAGUCAAGCAUUUGGGCAG .........(((((((((...((...((((((((..((((.....))))..(((........))))))))------((((....)))))))...))...)))))))))... ( -38.10) >DroPse_CAF1 32399 108 - 1 AGCCAAAUACCUGACUGUGACCCGUCGUCGCCGGCAGGCCGUCAAGGCCAACGGUCA---UGCCAAUGGCCAUGGCCUGCACACAAACGGGCACAGCAAGCACUUGGGCAA .(((......((..(((((.(((((........(((((((((...(((((..((...---..))..))))))))))))))......))))))))))..))......))).. ( -50.44) >DroMoj_CAF1 42578 102 - 1 GGCUAAAUAUCUGAAUGCAAAGCGUCGCCGCCGCGAGGCAGUCGAAGUCAAUGGCCA---UGCCAAUGGC------GUCUACACGAAUGGCAAAGUCAAGCAUUUAGGCAA .........(((((((((...((...((((((....))).((.((.((((.((((..---.)))).))))------.)).))......)))...))...)))))))))... ( -36.00) >DroAna_CAF1 29119 102 - 1 AGCCAAAUACCUGAGUGCCACCCGGCGUCGUCGCCAGGCUGUCAAGGCCAACGGACA---UGCCAACGGU------GUGCAAUCCAACGGGCACUCCAAGCACCUGGGGGA .(((........((.(((((((.((((....)))).(((((((..(.....).))))---.)))...)))------).))).)).....))).(((((......))))).. ( -36.82) >DroPer_CAF1 32838 108 - 1 AGCCAAAUACCUGACUGUGACCCGUCGUCGCCGGCAGGCCGUCAAGGCCAACGGUCA---UGCCAAUGGCCAUGGCCUGCACACAAACGGGCACAGCAAGCACUUGGGCAA .(((......((..(((((.(((((........(((((((((...(((((..((...---..))..))))))))))))))......))))))))))..))......))).. ( -50.44) >consensus AGCCAAAUACCUGAAUGCGACCCGUCGUCGCCGGCAGGCCGUCAAGGCCAACGGCCA___UGCCAACGGC______GUGCACACGAACGGGCACAGCAAGCACUUGGGCAA .........(((((((((...........((((...((((.....))))..))))......(((...))).............................)))))))))... (-18.80 = -18.92 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:00 2006