| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,003,440 – 10,003,549 |
| Length | 109 |
| Max. P | 0.625226 |

| Location | 10,003,440 – 10,003,549 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.38 |
| Mean single sequence MFE | -40.09 |
| Consensus MFE | -19.97 |
| Energy contribution | -19.67 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
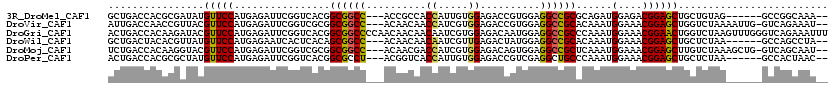
>3R_DroMel_CAF1 10003440 109 - 27905053 GCUGACCACGCGAUAUGUUCCAUGAGAUUCGGUCACGGCGGCC---ACCGCCACCAUUGUGGAGACCGUGGAGGCCGCGCAGAUGGAGACGGAGCUGCUGUAG------GCCGGCAAA-- (((..(((((.(.....(((((..(.....(((...(((((..---.)))))))).)..))))).)))))).((((((((((.((....))...)))).)).)------))))))...-- ( -44.60) >DroVir_CAF1 123909 114 - 1 AUUGACCAACCGUUACGUUCCAUGAGAUUCGGUCGCGGCGGCC---ACAACAACCAUCGUGGAGACCGUGGAGGCCGCACAAAUGGAAACGGAGCUGGUCUAAAAUUG-GUCAGAAAU-- .(((((((((((((.((((((((......(((((.(.((((((---((..........))))...)))).).))))).....)))).)))).))).))).......))-)))))....-- ( -40.11) >DroGri_CAF1 109012 120 - 1 ACUGACCACAAGAUACGUUCCAUGAGAUUCGGUCACGGCGGCCCCAACAACAACAAUCGUGGAGACAAUGGAGGCCGCCCAAAUGGAAACGGAACUGGUCUAAGUUUGGGUCAGAAAUUU .((((((.((((((..(((((...............((((((((((......((....))(....)..))).))))))).....(....))))))..)))).....))))))))...... ( -43.10) >DroWil_CAF1 92335 109 - 1 GCUGACUACACGUUAUGUUCCAUGAGAAUCACUCACAGCGGCC---ACAACAACAAUCGUUGAGACUAUGGAGGCCGCACAAAUGGAAACGGAGCUGCUCUAA------GCCAGCCUA-- ((((.((....((...(((((.((((.....))))..((((((---....((((....))))...(....).))))))......(....)))))).))....)------).))))...-- ( -32.80) >DroMoj_CAF1 112354 114 - 1 UCUGACCACAAGGUACGUUCCAUGAGAUUCGGUCGCGGCGGCC---ACAACGACCAUCGUGGAGACAGUGGAGGCCGCUCAAAUGGAAACGGAGCUUGUCUAAAGCUG-GUCAGCAAU-- .((((((........((((((((..(((...)))(((((..((---((.(((.....)))(....).))))..)))))....)))).)))).(((((.....))))))-)))))....-- ( -44.60) >DroPer_CAF1 89408 109 - 1 ACUGACCACGCGCUAUGUUCCAUGAGAUUCGGUCACGGCGCCU---ACGGUCACCAUUGUGGAGACCGUCGAGGCUGCCCAAAUGGAAACGGAGCUGCUCUAA------GCCACUAAC-- .........(((((.((((((((..(((...)))..(((((((---((((((.((.....)).))))))..)))).)))...)))).)))).))).)).....------.........-- ( -35.30) >consensus ACUGACCACACGAUACGUUCCAUGAGAUUCGGUCACGGCGGCC___ACAACAACCAUCGUGGAGACCGUGGAGGCCGCACAAAUGGAAACGGAGCUGCUCUAA______GCCAGCAAU__ ................(((((................((((((..........((.....))..........))))))......(....))))))......................... (-19.97 = -19.67 + -0.30)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:35 2006