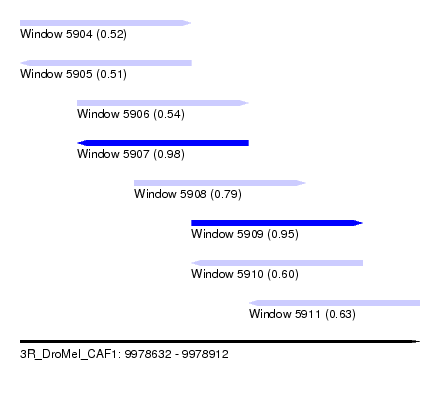
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,978,632 – 9,978,912 |
| Length | 280 |
| Max. P | 0.982922 |

| Location | 9,978,632 – 9,978,752 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.58 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -12.98 |
| Energy contribution | -15.30 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.45 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.520904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9978632 120 + 27905053 CGACGUCGAAUUGCAGAUGUAAAAUAAUCUGUGUGUUUGUGAGAUUUCAUGCAACUAAUCGGAUCCUCCGAUUAAUUGCAAAAUGUCCAGGAAUAUGUUCAUUACAUCCAUGUAAAUAUU .......((((.((((((........))))))((((((.((.(((....(((((.(((((((.....))))))).)))))....))))).)))))))))).(((((....)))))..... ( -34.00) >DroSec_CAF1 61052 120 + 1 CGUCGUCGAAUUGGUGGUGUAAAAUAAUCUGUGUAUUUGUGGAAUUUCAUGCAACUAAUCUGAUCCUCCGAUUAAUUGCAAAACGUCCAGGAAUAUGUUCAUCACAUCCAUGUAAAUAUU ...(((.((....((((((...........(..(((((.((((......(((((.(((((.(.....).))))).))))).....)))).)))))..).)))))).)).)))........ ( -26.30) >DroSim_CAF1 61019 120 + 1 CGUCGUCGAAUUGGAUGUGUAAAAUAAUCUGUGUAUUUGUGGGAUUUCAUGCAACUAAUCGGAUCCUCCGAUUAAUUGCAAAAUGUCCAGGAAUAUGUUCAUCACAUCCAUGUAAAUAUU ...........((((((((...........(..(((((.((((((((..(((((.(((((((.....))))))).))))))))).)))).)))))..)....)))))))).......... ( -35.66) >DroEre_CAF1 65559 103 + 1 UU---------------AUUAAGAUAAUACGUG--UCUUUGGGAUUUUAUGGAACUAAUCGGAUCCCCCGAGUAACUGCAAUAUUUCCAGGAAUAUGUUCAUAACAUCCAUAUAAAUAUU ..---------------...((((((.....))--))))..((((.(((((((.....((((.....)))).........((((((....)))))).))))))).))))........... ( -20.10) >DroYak_CAF1 63942 120 + 1 CGACGUCGAAUUGGUGGAGUAAAAUAGUACGAGUAUUUUUGGGAUUUCAUCGAACUAAUCAGAUCCUCCGAUUAAUUGCAAGAUGUCGUGGAAUAUGUUUAUCACAUCCAUAUAAAUAUU .((((((.((((((.(((......((((.(((..(((.....)))....))).))))......))).))))))........))))))(((((...(((.....))))))))......... ( -27.30) >consensus CGACGUCGAAUUGGAGGUGUAAAAUAAUCUGUGUAUUUGUGGGAUUUCAUGCAACUAAUCGGAUCCUCCGAUUAAUUGCAAAAUGUCCAGGAAUAUGUUCAUCACAUCCAUGUAAAUAUU ................................((((((.((((......(((((.(((((((.....))))))).))))).....)))).))))))........................ (-12.98 = -15.30 + 2.32)


| Location | 9,978,632 – 9,978,752 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.58 |
| Mean single sequence MFE | -24.37 |
| Consensus MFE | -17.24 |
| Energy contribution | -18.36 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9978632 120 - 27905053 AAUAUUUACAUGGAUGUAAUGAACAUAUUCCUGGACAUUUUGCAAUUAAUCGGAGGAUCCGAUUAGUUGCAUGAAAUCUCACAAACACACAGAUUAUUUUACAUCUGCAAUUCGACGUCG ............(((((...(((........((((.(((((((((((((((((.....)))))))))))))..)))).)).))......(((((........)))))...))).))))). ( -29.10) >DroSec_CAF1 61052 120 - 1 AAUAUUUACAUGGAUGUGAUGAACAUAUUCCUGGACGUUUUGCAAUUAAUCGGAGGAUCAGAUUAGUUGCAUGAAAUUCCACAAAUACACAGAUUAUUUUACACCACCAAUUCGACGACG ..........(((.(((((......((((..((((..((.(((((((((((.(.....).))))))))))).))...))))..))))...........)))))))).............. ( -22.83) >DroSim_CAF1 61019 120 - 1 AAUAUUUACAUGGAUGUGAUGAACAUAUUCCUGGACAUUUUGCAAUUAAUCGGAGGAUCCGAUUAGUUGCAUGAAAUCCCACAAAUACACAGAUUAUUUUACACAUCCAAUUCGACGACG ..........((((((((.((((.(((.((..(((..((.(((((((((((((.....))))))))))))).))..)))............)).)))))))))))))))........... ( -33.54) >DroEre_CAF1 65559 103 - 1 AAUAUUUAUAUGGAUGUUAUGAACAUAUUCCUGGAAAUAUUGCAGUUACUCGGGGGAUCCGAUUAGUUCCAUAAAAUCCCAAAGA--CACGUAUUAUCUUAAU---------------AA ...........((((.((((((((......(((.(.....).)))....((((.....))))...))).))))).))))......--................---------------.. ( -15.00) >DroYak_CAF1 63942 120 - 1 AAUAUUUAUAUGGAUGUGAUAAACAUAUUCCACGACAUCUUGCAAUUAAUCGGAGGAUCUGAUUAGUUCGAUGAAAUCCCAAAAAUACUCGUACUAUUUUACUCCACCAAUUCGACGUCG ..........((((((((.....)))).))))((((.(((((.((((((((((.....))))))))))))).)).......((((((.......))))))................)))) ( -21.40) >consensus AAUAUUUACAUGGAUGUGAUGAACAUAUUCCUGGACAUUUUGCAAUUAAUCGGAGGAUCCGAUUAGUUGCAUGAAAUCCCACAAAUACACAGAUUAUUUUACACCACCAAUUCGACGACG ...........(((((((.....)))))))..(((..((.(((((((((((((.....))))))))))))).))..)))......................................... (-17.24 = -18.36 + 1.12)



| Location | 9,978,672 – 9,978,792 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -30.32 |
| Consensus MFE | -24.06 |
| Energy contribution | -25.62 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9978672 120 + 27905053 GAGAUUUCAUGCAACUAAUCGGAUCCUCCGAUUAAUUGCAAAAUGUCCAGGAAUAUGUUCAUUACAUCCAUGUAAAUAUUUAGGGCAGCGAAUAUGUACUCCUCGGGACUAAUGGAGUAC .....(((.(((((.(((((((.....))))))).)))))...(((((..((((((...(((.......)))...)))))).)))))..)))...(((((((...........))))))) ( -35.00) >DroSec_CAF1 61092 120 + 1 GGAAUUUCAUGCAACUAAUCUGAUCCUCCGAUUAAUUGCAAAACGUCCAGGAAUAUGUUCAUCACAUCCAUGUAAAUAUUUAGGGCGGCGAAUAUGUACUCCUCGGGACUAAUGGAGUAC .....(((.(((((.(((((.(.....).))))).)))))...(((((..((((((...(((.......)))...)))))).)))))..)))...(((((((...........))))))) ( -30.40) >DroSim_CAF1 61059 120 + 1 GGGAUUUCAUGCAACUAAUCGGAUCCUCCGAUUAAUUGCAAAAUGUCCAGGAAUAUGUUCAUCACAUCCAUGUAAAUAUUUAGGGCGGCGAAUAUGUACUCCUCGGGACUAAUGGAGUAC ...(((((((((((.(((((((.....))))))).)))).....((((((((((((((((..(...(((.((........))))).)..))))))))..))))..))))..))))))).. ( -34.60) >DroEre_CAF1 65582 120 + 1 GGGAUUUUAUGGAACUAAUCGGAUCCCCCGAGUAACUGCAAUAUUUCCAGGAAUAUGUUCAUAACAUCCAUAUAAAUAUUUAGGGCCGCGAAUAUGUACUCCUCGGGACUUAUGGAGUAC .((((.(((((((.....((((.....)))).........((((((....)))))).))))))).))))......((((((........))))))(((((((...........))))))) ( -27.60) >DroYak_CAF1 63982 120 + 1 GGGAUUUCAUCGAACUAAUCAGAUCCUCCGAUUAAUUGCAAGAUGUCGUGGAAUAUGUUUAUCACAUCCAUAUAAAUAUUUAGGGCGGCGAAUAUGUACUCCUCGGGACUUAUGGAGUAC .....(((..((..((((...((((....))))........(((((.(((((.....))))).)))))...........))))..))..)))...(((((((...........))))))) ( -24.00) >consensus GGGAUUUCAUGCAACUAAUCGGAUCCUCCGAUUAAUUGCAAAAUGUCCAGGAAUAUGUUCAUCACAUCCAUGUAAAUAUUUAGGGCGGCGAAUAUGUACUCCUCGGGACUAAUGGAGUAC .....(((.(((((.(((((((.....))))))).)))))...(((((..((((((...(((.......)))...)))))).)))))..)))...(((((((...........))))))) (-24.06 = -25.62 + 1.56)

| Location | 9,978,672 – 9,978,792 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -30.74 |
| Consensus MFE | -27.34 |
| Energy contribution | -28.46 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9978672 120 - 27905053 GUACUCCAUUAGUCCCGAGGAGUACAUAUUCGCUGCCCUAAAUAUUUACAUGGAUGUAAUGAACAUAUUCCUGGACAUUUUGCAAUUAAUCGGAGGAUCCGAUUAGUUGCAUGAAAUCUC ...........((((..(((((((....((((.(((((.............))..))).))))..)))))))))))....(((((((((((((.....)))))))))))))......... ( -34.12) >DroSec_CAF1 61092 120 - 1 GUACUCCAUUAGUCCCGAGGAGUACAUAUUCGCCGCCCUAAAUAUUUACAUGGAUGUGAUGAACAUAUUCCUGGACGUUUUGCAAUUAAUCGGAGGAUCAGAUUAGUUGCAUGAAAUUCC (((((((...........)))))))...((((................((.(((((((.....))))))).)).......(((((((((((.(.....).)))))))))))))))..... ( -30.40) >DroSim_CAF1 61059 120 - 1 GUACUCCAUUAGUCCCGAGGAGUACAUAUUCGCCGCCCUAAAUAUUUACAUGGAUGUGAUGAACAUAUUCCUGGACAUUUUGCAAUUAAUCGGAGGAUCCGAUUAGUUGCAUGAAAUCCC (((((((...........)))))))..........................(((((((.....)))))))..(((..((.(((((((((((((.....))))))))))))).))..))). ( -36.80) >DroEre_CAF1 65582 120 - 1 GUACUCCAUAAGUCCCGAGGAGUACAUAUUCGCGGCCCUAAAUAUUUAUAUGGAUGUUAUGAACAUAUUCCUGGAAAUAUUGCAGUUACUCGGGGGAUCCGAUUAGUUCCAUAAAAUCCC (((((((...........)))))))..........................((((.((((((((......(((.(.....).)))....((((.....))))...))).))))).)))). ( -25.10) >DroYak_CAF1 63982 120 - 1 GUACUCCAUAAGUCCCGAGGAGUACAUAUUCGCCGCCCUAAAUAUUUAUAUGGAUGUGAUAAACAUAUUCCACGACAUCUUGCAAUUAAUCGGAGGAUCUGAUUAGUUCGAUGAAAUCCC (((((((...........))))))).........................((((((((.....)))).))))...((((....((((((((((.....)))))))))).))))....... ( -27.30) >consensus GUACUCCAUUAGUCCCGAGGAGUACAUAUUCGCCGCCCUAAAUAUUUACAUGGAUGUGAUGAACAUAUUCCUGGACAUUUUGCAAUUAAUCGGAGGAUCCGAUUAGUUGCAUGAAAUCCC (((((((...........)))))))..........................(((((((.....)))))))..(((..((.(((((((((((((.....))))))))))))).))..))). (-27.34 = -28.46 + 1.12)

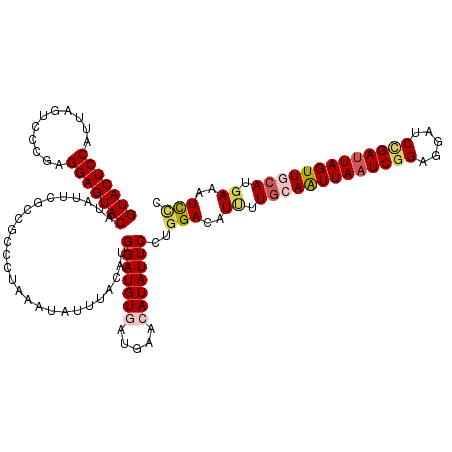
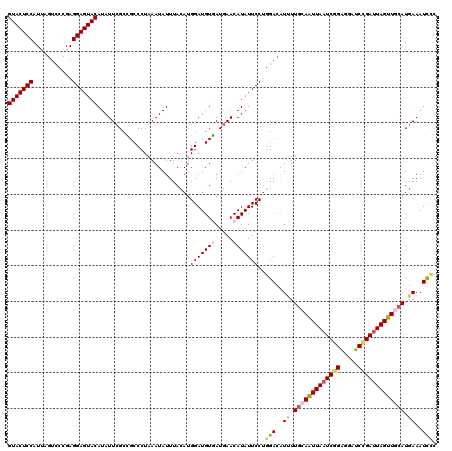
| Location | 9,978,712 – 9,978,832 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.08 |
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -24.68 |
| Energy contribution | -24.68 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9978712 120 + 27905053 AAAUGUCCAGGAAUAUGUUCAUUACAUCCAUGUAAAUAUUUAGGGCAGCGAAUAUGUACUCCUCGGGACUAAUGGAGUACCUAUGCUUGCCUCCCAUCAUCAAUUGUGUAUCGUAAACUA ..(((..(((((((((...(((.......)))...)))))).(((..(((((((.(((((((...........))))))).)))..))))..)))........)))..)))......... ( -23.90) >DroSec_CAF1 61132 120 + 1 AAACGUCCAGGAAUAUGUUCAUCACAUCCAUGUAAAUAUUUAGGGCGGCGAAUAUGUACUCCUCGGGACUAAUGGAGUACCUAUGCUUGCCUCCCAUCAUCAAUUGUGUAUCGUAAACCA .........((..((((.....((((...((((((....)))(((.((((((((.(((((((...........))))))).)))..))))).)))..)))....))))...))))..)). ( -29.30) >DroSim_CAF1 61099 120 + 1 AAAUGUCCAGGAAUAUGUUCAUCACAUCCAUGUAAAUAUUUAGGGCGGCGAAUAUGUACUCCUCGGGACUAAUGGAGUACCUAUGCUUGCCUCCCAUCAUCAAUUGUGUAUCGUAAACCA .........((..((((.....((((...((((((....)))(((.((((((((.(((((((...........))))))).)))..))))).)))..)))....))))...))))..)). ( -29.30) >DroEre_CAF1 65622 120 + 1 AUAUUUCCAGGAAUAUGUUCAUAACAUCCAUAUAAAUAUUUAGGGCCGCGAAUAUGUACUCCUCGGGACUUAUGGAGUACCUAUGCUUGCCUCCCAUCAUCAAUUGGGUAUCGUGAACCA ((((((....))))))(((((..(.(((((............(((..(((((((.(((((((...........))))))).)))..))))..))).........))))).)..))))).. ( -30.00) >DroYak_CAF1 64022 120 + 1 AGAUGUCGUGGAAUAUGUUUAUCACAUCCAUAUAAAUAUUUAGGGCGGCGAAUAUGUACUCCUCGGGACUUAUGGAGUACCUAUGCUUGCCUCCCAACAUCAAUUGGGUAUCGUAAACCA .(((((.(((((...(((.....))))))))...........(((.((((((((.(((((((...........))))))).)))..))))).))).))))).....(((.......))). ( -35.40) >consensus AAAUGUCCAGGAAUAUGUUCAUCACAUCCAUGUAAAUAUUUAGGGCGGCGAAUAUGUACUCCUCGGGACUAAUGGAGUACCUAUGCUUGCCUCCCAUCAUCAAUUGUGUAUCGUAAACCA .........((.(((((...........))))).........(((.((((((((.(((((((...........))))))).)))..))))).)))......................)). (-24.68 = -24.68 + -0.00)

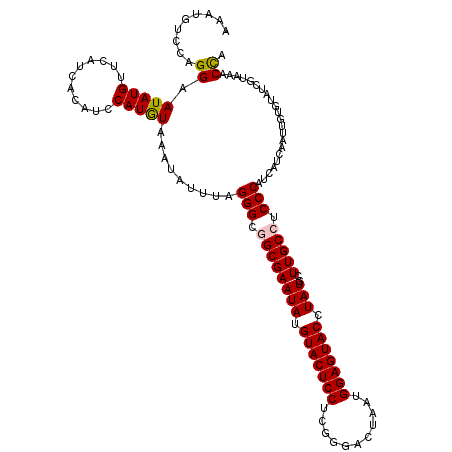
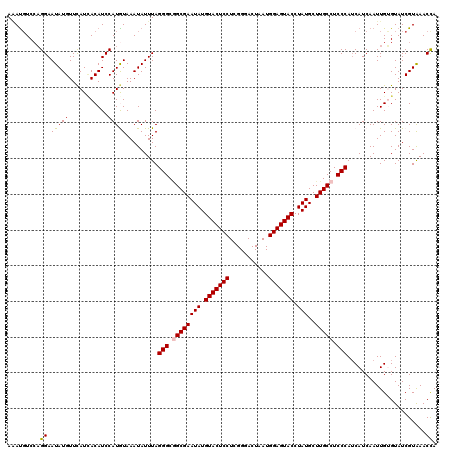
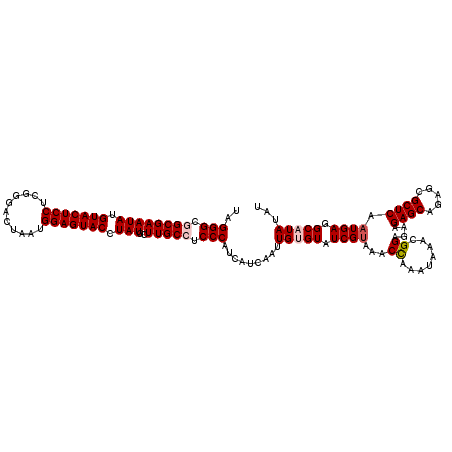
| Location | 9,978,752 – 9,978,872 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.92 |
| Mean single sequence MFE | -34.68 |
| Consensus MFE | -32.68 |
| Energy contribution | -33.52 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.949837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9978752 120 + 27905053 UAGGGCAGCGAAUAUGUACUCCUCGGGACUAAUGGAGUACCUAUGCUUGCCUCCCAUCAUCAAUUGUGUAUCGUAAACUAAAUAAACGGAGAAGAGCAGAGCGCUCAAUGAGGCAUAUAU ..(((..(((((((.(((((((...........))))))).)))..))))..))).........(((((.((((...((........))....((((.....)))).)))).)))))... ( -30.70) >DroSec_CAF1 61172 120 + 1 UAGGGCGGCGAAUAUGUACUCCUCGGGACUAAUGGAGUACCUAUGCUUGCCUCCCAUCAUCAAUUGUGUAUCGUAAACCAAAUAAACGGAGAAGAGCAGAGCGCUCAAUGAGGCAUAUAU ..(((.((((((((.(((((((...........))))))).)))..))))).))).........(((((.((((...((........))....((((.....)))).)))).)))))... ( -37.50) >DroSim_CAF1 61139 120 + 1 UAGGGCGGCGAAUAUGUACUCCUCGGGACUAAUGGAGUACCUAUGCUUGCCUCCCAUCAUCAAUUGUGUAUCGUAAACCAAAUAAACGGAGAAGAGCAGAGCGCUCAAUGAGGCAUAUAU ..(((.((((((((.(((((((...........))))))).)))..))))).))).........(((((.((((...((........))....((((.....)))).)))).)))))... ( -37.50) >DroEre_CAF1 65662 120 + 1 UAGGGCCGCGAAUAUGUACUCCUCGGGACUUAUGGAGUACCUAUGCUUGCCUCCCAUCAUCAAUUGGGUAUCGUGAACCAAAUAAAUGGAAAAGAGCAGGGCGCUCAAUGAGGCAUAGAU ....((((((((((.(((((((...........))))))).)))..))))..((((........))))..((((...(((......)))....((((.....)))).)))))))...... ( -33.10) >DroYak_CAF1 64062 120 + 1 UAGGGCGGCGAAUAUGUACUCCUCGGGACUUAUGGAGUACCUAUGCUUGCCUCCCAACAUCAAUUGGGUAUCGUAAACCAAGUAAACGGAAAAGAGCAGGGCGCUCAUUGAGGCAUAGAU ..(((.((((((((.(((((((...........))))))).)))..))))).)))....((((((((..........))))............((((.....)))).))))......... ( -34.60) >consensus UAGGGCGGCGAAUAUGUACUCCUCGGGACUAAUGGAGUACCUAUGCUUGCCUCCCAUCAUCAAUUGUGUAUCGUAAACCAAAUAAACGGAGAAGAGCAGAGCGCUCAAUGAGGCAUAUAU ..(((.((((((((.(((((((...........))))))).)))..))))).))).........(((((.((((...((........))....((((.....)))).)))).)))))... (-32.68 = -33.52 + 0.84)

| Location | 9,978,752 – 9,978,872 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.92 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -29.96 |
| Energy contribution | -29.84 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9978752 120 - 27905053 AUAUAUGCCUCAUUGAGCGCUCUGCUCUUCUCCGUUUAUUUAGUUUACGAUACACAAUUGAUGAUGGGAGGCAAGCAUAGGUACUCCAUUAGUCCCGAGGAGUACAUAUUCGCUGCCCUA .....((((((...((((.....))))....(((((...((((((..........)))))).)))))))))))((((((.(((((((...........))))))).)))..)))...... ( -32.90) >DroSec_CAF1 61172 120 - 1 AUAUAUGCCUCAUUGAGCGCUCUGCUCUUCUCCGUUUAUUUGGUUUACGAUACACAAUUGAUGAUGGGAGGCAAGCAUAGGUACUCCAUUAGUCCCGAGGAGUACAUAUUCGCCGCCCUA .....((((((...((((.....))))....(((((...(((((.......)).))).....))))))))))).(((((.(((((((...........))))))).)))..))....... ( -31.50) >DroSim_CAF1 61139 120 - 1 AUAUAUGCCUCAUUGAGCGCUCUGCUCUUCUCCGUUUAUUUGGUUUACGAUACACAAUUGAUGAUGGGAGGCAAGCAUAGGUACUCCAUUAGUCCCGAGGAGUACAUAUUCGCCGCCCUA .....((((((...((((.....))))....(((((...(((((.......)).))).....))))))))))).(((((.(((((((...........))))))).)))..))....... ( -31.50) >DroEre_CAF1 65662 120 - 1 AUCUAUGCCUCAUUGAGCGCCCUGCUCUUUUCCAUUUAUUUGGUUCACGAUACCCAAUUGAUGAUGGGAGGCAAGCAUAGGUACUCCAUAAGUCCCGAGGAGUACAUAUUCGCGGCCCUA .....((((((...((((.....))))....(((((.(((.(((.......))).)))....))))))))))).(((((.(((((((...........))))))).)))..))....... ( -34.50) >DroYak_CAF1 64062 120 - 1 AUCUAUGCCUCAAUGAGCGCCCUGCUCUUUUCCGUUUACUUGGUUUACGAUACCCAAUUGAUGUUGGGAGGCAAGCAUAGGUACUCCAUAAGUCCCGAGGAGUACAUAUUCGCCGCCCUA .....((((((...((((.....)))).....(((..((...))..)))....(((((....))))))))))).(((((.(((((((...........))))))).)))..))....... ( -31.90) >consensus AUAUAUGCCUCAUUGAGCGCUCUGCUCUUCUCCGUUUAUUUGGUUUACGAUACACAAUUGAUGAUGGGAGGCAAGCAUAGGUACUCCAUUAGUCCCGAGGAGUACAUAUUCGCCGCCCUA .....((((((...((((.....))))....(((((...((((((..........)))))).))))))))))).(((((.(((((((...........))))))).)))..))....... (-29.96 = -29.84 + -0.12)


| Location | 9,978,792 – 9,978,912 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.67 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -29.64 |
| Energy contribution | -29.96 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
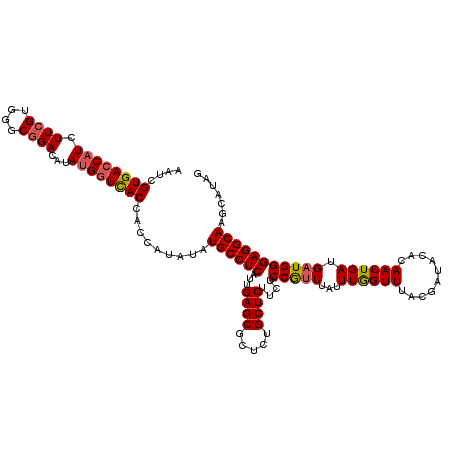
>3R_DroMel_CAF1 9978792 120 - 27905053 AAUUGUGACCAUCUUCGUGGGCGGACAUAUGGUCACCACCAUAUAUGCCUCAUUGAGCGCUCUGCUCUUCUCCGUUUAUUUAGUUUACGAUACACAAUUGAUGAUGGGAGGCAAGCAUAG ....((((((((.((((....))))...)))))))).........((((((...((((.....))))....(((((...((((((..........)))))).)))))))))))....... ( -34.40) >DroSec_CAF1 61212 120 - 1 AAUCGUGACCAUCUUCGUGGGCGGACAUAUGGUCACCACCAUAUAUGCCUCAUUGAGCGCUCUGCUCUUCUCCGUUUAUUUGGUUUACGAUACACAAUUGAUGAUGGGAGGCAAGCAUAG ....((((((((.((((....))))...)))))))).........((((((...((((.....))))....(((((...(((((.......)).))).....)))))))))))....... ( -34.90) >DroSim_CAF1 61179 120 - 1 AAUCGUGACCAUCUUCGUGGGCGGACAUAUGGUCACCACCAUAUAUGCCUCAUUGAGCGCUCUGCUCUUCUCCGUUUAUUUGGUUUACGAUACACAAUUGAUGAUGGGAGGCAAGCAUAG ....((((((((.((((....))))...)))))))).........((((((...((((.....))))....(((((...(((((.......)).))).....)))))))))))....... ( -34.90) >DroEre_CAF1 65702 120 - 1 AACCGUGAGCCUCUUAGUGGGCGGACAUAUGGUCACCACCAUCUAUGCCUCAUUGAGCGCCCUGCUCUUUUCCAUUUAUUUGGUUCACGAUACCCAAUUGAUGAUGGGAGGCAAGCAUAG ...((((((((....((((((.((.((((((((....))))..)))))).....((((.....))))...)))))).....))))))))...((((........))))............ ( -36.00) >DroYak_CAF1 64102 120 - 1 CAUCGUGACCAUCUUCGUGGGCGGACAUAUGGUUACCACCAUCUAUGCCUCAAUGAGCGCCCUGCUCUUUUCCGUUUACUUGGUUUACGAUACCCAAUUGAUGUUGGGAGGCAAGCAUAG .((((((((((.....(((((((((((((((((....))))..)))).......((((.....))))...))))))))).))).))))))).((((((....))))))............ ( -37.20) >consensus AAUCGUGACCAUCUUCGUGGGCGGACAUAUGGUCACCACCAUAUAUGCCUCAUUGAGCGCUCUGCUCUUCUCCGUUUAUUUGGUUUACGAUACACAAUUGAUGAUGGGAGGCAAGCAUAG ....((((((((.((((....))))...)))))))).........((((((...((((.....))))....(((((...((((((..........)))))).)))))))))))....... (-29.64 = -29.96 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:18 2006