
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,915,863 – 9,915,996 |
| Length | 133 |
| Max. P | 0.853759 |

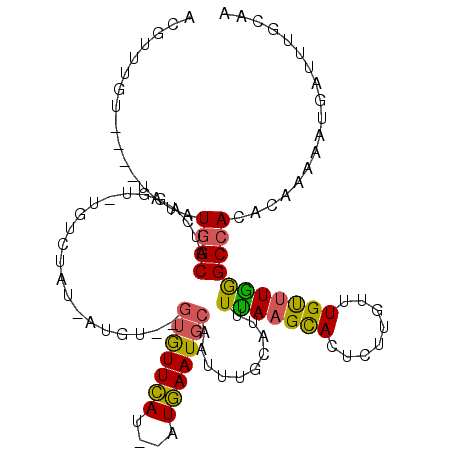
| Location | 9,915,863 – 9,915,969 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.60 |
| Mean single sequence MFE | -25.75 |
| Consensus MFE | -13.32 |
| Energy contribution | -12.80 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9915863 106 + 27905053 ACGUUUGU----UAUAUGGCAUCAGGU-UGUCUAU-AUGUGUGUGUUCAU--AUGAAUGCAAUUUGCAUUUUAAGCACCCUUGUUUGUUUGAGCCACACAAAAAUGAUUUGCAA ..((..((----(((..((((......-))))...-.((((((.(((((.--..((((((.....))))))((((((....))))))..)))))))))))...)))))..)).. ( -28.00) >DroPse_CAF1 143325 102 + 1 AUGGGUAA----UGAUUUGCUUAUGA------UAUAAUCUG-CUAUUGACUUGUCAAUGCAAUUUGCAUUUUAAGUA-UCGAAUGUGUCUAAGCCAAACAGAAAUGAUUGGCAA ((((((((----....))))))))((------((((.((((-(.(((((....))))))))...(((.......)))-..)).))))))...(((((.((....)).))))).. ( -26.90) >DroSec_CAF1 118783 104 + 1 ACGUAUGU----UAUAUGGCAUCUGGU-UGUCUAU-AUGU--GUGUUCAU--UUGAAUGCAAUUUGCAUUUUAAGCACUCUUGUUUGUUUGGGCCACACAAAAAUGAUUUGCUA ..(((.((----(((..((((......-))))...-.(((--(((..(..--..((((((.....))))))((((((........)))))))..))))))...))))).))).. ( -23.90) >DroSim_CAF1 122812 104 + 1 ACGCUUGU----UAUAUGGCAUCAGGU-UGUCUAU-AUGU--GUGUUCAU--UUGAAUGCAAUUUGCAUUUUAAGCACUCUUGUUUGUUUGGGCCACACAAAAAUGAUUUGCAA ..((..((----(((..((((......-))))...-.(((--(((..(..--..((((((.....))))))((((((........)))))))..))))))...)))))..)).. ( -25.70) >DroEre_CAF1 124214 104 + 1 UCGUUUGU----UAUAUGGCAUCAGGUUUGUCUAU-AUGU--GAGUUCAU--AUGAAUGCA-UUUGCAUUUUAAGCACUCUUGUUUGUUUGGGCCACACAAAAAUGAUUUGUAA ((((((..----((((.((((.......)))))))-)(((--(.(((((.--..((((((.-...))))))((((((....))))))..)))))))))...))))))....... ( -21.90) >DroYak_CAF1 123244 109 + 1 UAGUUUGUGUACUGUAUGGCAGCAAGUUUUUCUAC-AUGU--GUGUUCAU--AUGAAUGCAAUUUGCAUUUCAAGCACUCUUGUUUGCUUGGGCCACACAAAAAUGAUUUGCAA ((((......)))).......(((((((.......-.(((--(((..(..--..((((((.....))))))((((((........)))))))..)))))).....))))))).. ( -28.12) >consensus ACGUUUGU____UAUAUGGCAUCAGGU_UGUCUAU_AUGU__GUGUUCAU__AUGAAUGCAAUUUGCAUUUUAAGCACUCUUGUUUGUUUGGGCCACACAAAAAUGAUUUGCAA ................((((......................(((((((....)))))))..........(((((((........))))))))))).................. (-13.32 = -12.80 + -0.52)

| Location | 9,915,863 – 9,915,969 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.60 |
| Mean single sequence MFE | -19.74 |
| Consensus MFE | -12.27 |
| Energy contribution | -12.06 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9915863 106 - 27905053 UUGCAAAUCAUUUUUGUGUGGCUCAAACAAACAAGGGUGCUUAAAAUGCAAAUUGCAUUCAU--AUGAACACACACAU-AUAGACA-ACCUGAUGCCAUAUA----ACAAACGU .............((((((((((((.........((((((..............))))))((--(((........)))-)).....-...))).))))))))----)....... ( -20.84) >DroPse_CAF1 143325 102 - 1 UUGCCAAUCAUUUCUGUUUGGCUUAGACACAUUCGA-UACUUAAAAUGCAAAUUGCAUUGACAAGUCAAUAG-CAGAUUAUA------UCAUAAGCAAAUCA----UUACCCAU ..(((((.((....)).)))))............((-(.((((..(((.((.(((((((((....))))).)-))).)).))------)..))))...))).----........ ( -17.20) >DroSec_CAF1 118783 104 - 1 UAGCAAAUCAUUUUUGUGUGGCCCAAACAAACAAGAGUGCUUAAAAUGCAAAUUGCAUUCAA--AUGAACAC--ACAU-AUAGACA-ACCAGAUGCCAUAUA----ACAUACGU .............(((((((((............((((((..............))))))..--(((.....--.)))-.......-.......))))))))----)....... ( -17.24) >DroSim_CAF1 122812 104 - 1 UUGCAAAUCAUUUUUGUGUGGCCCAAACAAACAAGAGUGCUUAAAAUGCAAAUUGCAUUCAA--AUGAACAC--ACAU-AUAGACA-ACCUGAUGCCAUAUA----ACAAGCGU .............(((((((((............((((((..............))))))..--........--....-.(((...-..)))..))))))))----)....... ( -17.44) >DroEre_CAF1 124214 104 - 1 UUACAAAUCAUUUUUGUGUGGCCCAAACAAACAAGAGUGCUUAAAAUGCAAA-UGCAUUCAU--AUGAACUC--ACAU-AUAGACAAACCUGAUGCCAUAUA----ACAAACGA .............(((((((((............((((((............-.))))))((--(((.....--.)))-)).............))))))))----)....... ( -19.72) >DroYak_CAF1 123244 109 - 1 UUGCAAAUCAUUUUUGUGUGGCCCAAGCAAACAAGAGUGCUUGAAAUGCAAAUUGCAUUCAU--AUGAACAC--ACAU-GUAGAAAAACUUGCUGCCAUACAGUACACAAACUA .............(((((((((.((((((........))))))....((((.((...(((((--(((.....--.)))-)).))).)).)))).)))))))))........... ( -26.00) >consensus UUGCAAAUCAUUUUUGUGUGGCCCAAACAAACAAGAGUGCUUAAAAUGCAAAUUGCAUUCAU__AUGAACAC__ACAU_AUAGACA_ACCUGAUGCCAUAUA____ACAAACGU ..............((((((((............((((((..............))))))....(((........)))................))))))))............ (-12.27 = -12.06 + -0.22)


| Location | 9,915,892 – 9,915,996 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.22 |
| Mean single sequence MFE | -22.73 |
| Consensus MFE | -14.55 |
| Energy contribution | -13.55 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9915892 104 - 27905053 CGCGAAGUCU-------------GGAGACCAUAAAAAGUUUUGCAAAUCAUUUUUGUGUGGCUCAAACAAACAAGGGUGCUUAAAAUGCAAAUUGCAUUCAU--AUGAACACACACAU-A .((((((.((-------------(....).......))))))))..........((((((((((..........))))......(((((.....)))))...--.......)))))).-. ( -18.61) >DroPse_CAF1 143349 118 - 1 CAUAAAGUCUCAAGAGCCAACCAGAAGCUCGUAGAAAAUGUUGCCAAUCAUUUCUGUUUGGCUUAGACACAUUCGA-UACUUAAAAUGCAAAUUGCAUUGACAAGUCAAUAG-CAGAUUA ......((((...(((((((.((((((...((((......))))......)))))).)))))))))))........-...............(((((((((....))))).)-))).... ( -26.30) >DroSec_CAF1 118812 102 - 1 CGCGAAGUCU-------------GGAGCCCAUAAAAAGUUUAGCAAAUCAUUUUUGUGUGGCCCAAACAAACAAGAGUGCUUAAAAUGCAAAUUGCAUUCAA--AUGAACAC--ACAU-A ......((.(-------------((.(((((((((((.............)))))))).)))))).))......((((((..............))))))..--........--....-. ( -22.86) >DroSim_CAF1 122841 102 - 1 CGCGAAGUCU-------------GGAGUCCAUAAAAAGUUUUGCAAAUCAUUUUUGUGUGGCCCAAACAAACAAGAGUGCUUAAAAUGCAAAUUGCAUUCAA--AUGAACAC--ACAU-A ......((.(-------------((.(((((((((((.............)))))))).)))))).))......((((((..............))))))..--........--....-. ( -20.16) >DroEre_CAF1 124244 101 - 1 UGCAAAGUCU-------------GGAGCCCAUAAAAAGCUUUACAAAUCAUUUUUGUGUGGCCCAAACAAACAAGAGUGCUUAAAAUGCAAA-UGCAUUCAU--AUGAACUC--ACAU-A ......((.(-------------((.(((((((((((.............)))))))).)))))).))......((((......(((((...-.)))))...--....))))--....-. ( -24.64) >DroYak_CAF1 123278 102 - 1 CGCAAAGUCU-------------GGAGCCCAUAAAAAGUUUUGCAAAUCAUUUUUGUGUGGCCCAAGCAAACAAGAGUGCUUGAAAUGCAAAUUGCAUUCAU--AUGAACAC--ACAU-G .((((((.((-------------.............))))))))..........((((((...((((((........)))))).(((((.....)))))...--.....)))--))).-. ( -23.82) >consensus CGCAAAGUCU_____________GGAGCCCAUAAAAAGUUUUGCAAAUCAUUUUUGUGUGGCCCAAACAAACAAGAGUGCUUAAAAUGCAAAUUGCAUUCAU__AUGAACAC__ACAU_A .((((..................((.(((((((((((.............)))))))).))))).............(((.......)))..)))).((((....))))........... (-14.55 = -13.55 + -0.99)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:52 2006