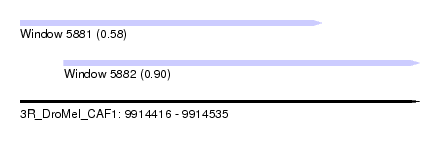
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,914,416 – 9,914,535 |
| Length | 119 |
| Max. P | 0.897138 |

| Location | 9,914,416 – 9,914,506 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -19.95 |
| Consensus MFE | -13.65 |
| Energy contribution | -14.27 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9914416 90 + 27905053 CACAACCGCAAAGUAUUCAAAUCCAAUCGAAUCCACAGCCUCGACAUCAU---AUCGAAAUUGCUGUGUCAGUAUCGAUUGUGACGCAUGAUU .......((...(....)...(((((((((...((((((.((((......---.))))....))))))......))))))).)).))...... ( -18.80) >DroSec_CAF1 117289 90 + 1 CACAACCGCAAAGUACUCAAAUCCAAUCGAAUCCACAGCCUCGUCAUCAU---AUCGAAAUUGCUGUGUCCGUAUCGAUUGUGAUGCUUGAUU ................((((((((((((((...((((((....((.....---...))....))))))......))))))).)))..)))).. ( -19.80) >DroSim_CAF1 121339 90 + 1 CACAACCGCAAAGUACUCAAAUCCAAUCGAAUACACAGUCUCGUCAUCAU---AUCGAAAUUGCUGUGUCCGUAUCGAUUGUGAUGCAUAAUU .......((...(....)..((((((((((..(((((((....((.....---...))....))))))).....))))))).)))))...... ( -18.90) >DroEre_CAF1 118375 92 + 1 CACAACUGCAGAGUACUCAUAUCCUAUUGAAUCCACAGCCUCGUCAUCAUCAAGUCGAAAUUGCUGUGUCCGUAGCGAUUGUGCUGCAUAAU- ......((((..((((....(((((((.((...((((((.(((.(........).)))....)))))))).)))).))).))))))))....- ( -22.30) >consensus CACAACCGCAAAGUACUCAAAUCCAAUCGAAUCCACAGCCUCGUCAUCAU___AUCGAAAUUGCUGUGUCCGUAUCGAUUGUGAUGCAUAAUU .......((...(....)..((((((((((...((((((.(((............)))....))))))......))))))).)))))...... (-13.65 = -14.27 + 0.63)

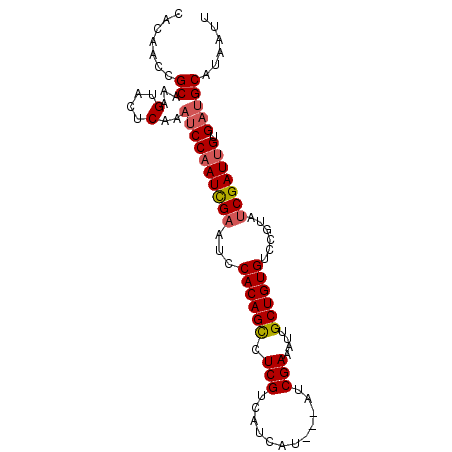
| Location | 9,914,429 – 9,914,535 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.71 |
| Mean single sequence MFE | -23.25 |
| Consensus MFE | -13.48 |
| Energy contribution | -14.10 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9914429 106 + 27905053 UAUUCAAAUCCAAUCGAAUCCACAGCCUCGACAUCAU---AUCGAAAUUGCUGUGUCAGUAUCGAUUGUGACGCAUGAUUUAUAAUAUC----GCAGUG-UU------AUCAAAGAUAAA .......((((((((((...((((((.((((......---.))))....))))))......)))))))((((((.((((.......)))----)..)))-))------).....)))... ( -22.90) >DroSec_CAF1 117302 114 + 1 UACUCAAAUCCAAUCGAAUCCACAGCCUCGUCAUCAU---AUCGAAAUUGCUGUGUCCGUAUCGAUUGUGAUGCUUGAUU---AAUAUCGAGCGCAGUGAUUAUCAUUAUCAAAGAUGAA (((((..((((((((((...((((((....((.....---...))....))))))......))))))).)))(((((((.---...)))))))).))))....(((((......))))). ( -31.30) >DroSim_CAF1 121352 104 + 1 UACUCAAAUCCAAUCGAAUACACAGUCUCGUCAUCAU---AUCGAAAUUGCUGUGUCCGUAUCGAUUGUGAUGCAUAAUU---AAUAUC----GCAGUGAUU------AUCAAAGAUGAA .......((((((((((..(((((((....((.....---...))....))))))).....))))))).)))........---..((((----....((...------..))..)))).. ( -20.80) >DroEre_CAF1 118388 85 + 1 UACUCAUAUCCUAUUGAAUCCACAGCCUCGUCAUCAUCAAGUCGAAAUUGCUGUGUCCGUAGCGAUUGUGCUGCAUAAU----AAUA-------------------------------CA ...........(((((....((((((.(((.(........).)))....))))))...((((((....)))))).))))----)...-------------------------------.. ( -18.00) >consensus UACUCAAAUCCAAUCGAAUCCACAGCCUCGUCAUCAU___AUCGAAAUUGCUGUGUCCGUAUCGAUUGUGAUGCAUAAUU___AAUAUC____GCAGUG_UU______AUCAAAGAUGAA .......((((((((((...((((((.(((............)))....))))))......))))))).)))................................................ (-13.48 = -14.10 + 0.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:49 2006