
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
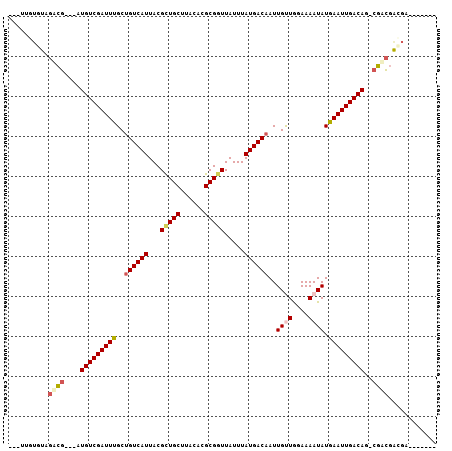
| Location | 9,904,664 – 9,904,757 |
| Length | 93 |
| Max. P | 0.599658 |

| Location | 9,904,664 – 9,904,757 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 82.63 |
| Mean single sequence MFE | -26.53 |
| Consensus MFE | -19.77 |
| Energy contribution | -21.58 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
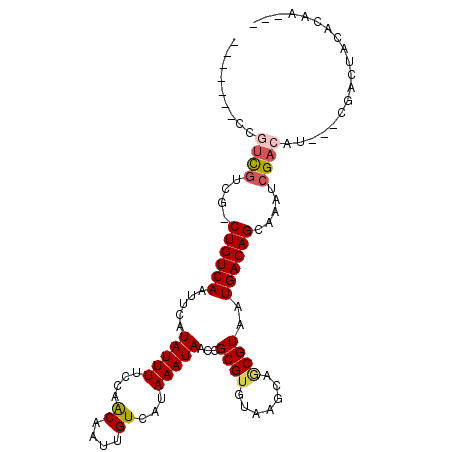
>3R_DroMel_CAF1 9904664 93 + 27905053 ---UUGUGUAGACG---AUGUCGAUUCGCUGUCAUUACGCUGCUUACACGCGGUUAUUUAUGACAAUUGUUGGAAAAUAUGAAUUGACAG-CGACGACGG------- ---((((....)))---)(((((..(((((((((....(((((......)))))(((((.((((....))))..))))).....))))))-)))))))).------- ( -27.30) >DroPse_CAF1 128588 98 + 1 UGUUGGUGUAGUCG--AAUGUCGAUUUGCUGUCAUUACGCUGCUUACACGCGGUUAUUUAUGACAAUUGCUGGAAAAUAUGAAUUGACAGGCGACGACGA------- ......(((.((((--..(((((((((((((((((...(((((......))))).....))))))...))(....)....)))))))))..)))).))).------- ( -28.90) >DroWil_CAF1 136028 97 + 1 ---GAAAAUAGAAUGGCUUGUCGAUUUGCUGUCAUUACGUUGCUUA--CGCGUUUAUUUAUGACUAUUGUUAUAAAAUAUGAAUUGACAG-CUUAAACAGCGA---- ---............(((.((..(...(((((((...(((.((...--.)).....((((((((....))))))))..)))...))))))-).)..)))))..---- ( -21.30) >DroYak_CAF1 111754 100 + 1 ---UUGUGUAGACG---AUGUCGAUUCGCUGUCAUUACGCUGCUUACACGCGGUUAUUUAUGACAAUUGUUGGAAAAUAUGAAUUGACAG-CGACGAUGGCGAUGGC ---.........((---..((((..(((((((((....(((((......)))))(((((.((((....))))..))))).....))))))-)))))))..))..... ( -27.40) >DroAna_CAF1 108095 94 + 1 ---CUGUGUAGUCG---AUGUCGAUUUGCUGUCAUUACGUUGCUUACACGCGGUUAUUUAUGACAAUUGUUGGAAAAUAUGAAUUGACAGUCGACGGGGA------- ---.......((((---((((((((((..((((((....((((......))))......))))))..((((....)))).))))))))).))))).....------- ( -25.40) >DroPer_CAF1 130251 98 + 1 UGUUGGUGUAGUCG--AAUGUCGAUUUGCUGUCAUUACGCUGCUUACACGCGGUUAUUUAUGACAAUUGCUGGAAAAUAUGAAUUGACAGGCGACGACGA------- ......(((.((((--..(((((((((((((((((...(((((......))))).....))))))...))(....)....)))))))))..)))).))).------- ( -28.90) >consensus ___UUGUGUAGACG___AUGUCGAUUUGCUGUCAUUACGCUGCUUACACGCGGUUAUUUAUGACAAUUGUUGGAAAAUAUGAAUUGACAG_CGACGACGA_______ ..........((((....(((((((((..((((((...(((((......))))).....))))))..((((....)))).)))))))))..))))............ (-19.77 = -21.58 + 1.81)


| Location | 9,904,664 – 9,904,757 |
|---|---|
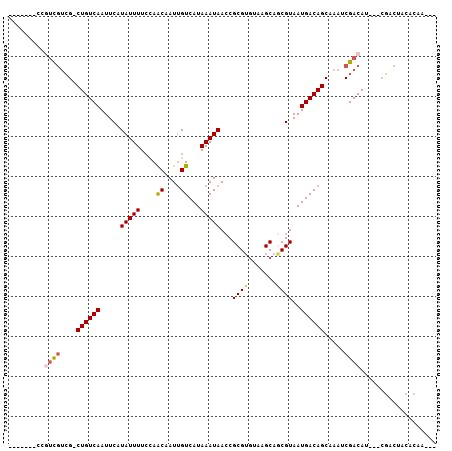
| Length | 93 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 82.63 |
| Mean single sequence MFE | -19.72 |
| Consensus MFE | -12.80 |
| Energy contribution | -13.17 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9904664 93 - 27905053 -------CCGUCGUCG-CUGUCAAUUCAUAUUUUCCAACAAUUGUCAUAAAUAACCGCGUGUAAGCAGCGUAAUGACAGCGAAUCGACAU---CGUCUACACAA--- -------..(((((((-((((((.....(((((....((....))...)))))..(((.((....)))))...)))))))))..))))..---...........--- ( -24.30) >DroPse_CAF1 128588 98 - 1 -------UCGUCGUCGCCUGUCAAUUCAUAUUUUCCAGCAAUUGUCAUAAAUAACCGCGUGUAAGCAGCGUAAUGACAGCAAAUCGACAUU--CGACUACACCAACA -------..((.((((..(((((((....))).....((...((((((.......(((.((....)))))..)))))))).....))))..--)))).))....... ( -18.40) >DroWil_CAF1 136028 97 - 1 ----UCGCUGUUUAAG-CUGUCAAUUCAUAUUUUAUAACAAUAGUCAUAAAUAAACGCG--UAAGCAACGUAAUGACAGCAAAUCGACAAGCCAUUCUAUUUUC--- ----..((((((...(-((((((........(((((.((....)).))))).....((.--...)).......))))))).....))).)))............--- ( -19.60) >DroYak_CAF1 111754 100 - 1 GCCAUCGCCAUCGUCG-CUGUCAAUUCAUAUUUUCCAACAAUUGUCAUAAAUAACCGCGUGUAAGCAGCGUAAUGACAGCGAAUCGACAU---CGUCUACACAA--- .....((...((((((-((((((.....(((((....((....))...)))))..(((.((....)))))...)))))))))..)))...---)).........--- ( -20.50) >DroAna_CAF1 108095 94 - 1 -------UCCCCGUCGACUGUCAAUUCAUAUUUUCCAACAAUUGUCAUAAAUAACCGCGUGUAAGCAACGUAAUGACAGCAAAUCGACAU---CGACUACACAG--- -------.....(((((((((((.....(((((....((....))...)))))...((((.......))))..))))))....)))))..---...........--- ( -17.10) >DroPer_CAF1 130251 98 - 1 -------UCGUCGUCGCCUGUCAAUUCAUAUUUUCCAGCAAUUGUCAUAAAUAACCGCGUGUAAGCAGCGUAAUGACAGCAAAUCGACAUU--CGACUACACCAACA -------..((.((((..(((((((....))).....((...((((((.......(((.((....)))))..)))))))).....))))..--)))).))....... ( -18.40) >consensus _______CCGUCGUCG_CUGUCAAUUCAUAUUUUCCAACAAUUGUCAUAAAUAACCGCGUGUAAGCAGCGUAAUGACAGCAAAUCGACAU___CGACUACACAA___ .........((((....((((((.....(((((....((....))...)))))...((((.......))))..)))))).....))))................... (-12.80 = -13.17 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:44 2006