
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,785,381 – 9,785,482 |
| Length | 101 |
| Max. P | 0.694341 |

| Location | 9,785,381 – 9,785,482 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 75.09 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -21.77 |
| Energy contribution | -22.38 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.10 |
| Mean z-score | -0.83 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
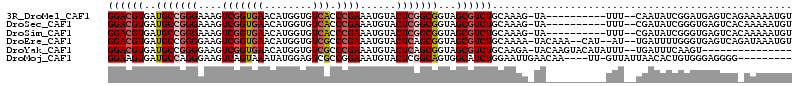
>3R_DroMel_CAF1 9785381 101 - 27905053 GGACGUGAUGCCGGGAAAGUCGGUGAACAUGGUGUCACCCGAAAUGUACUCGGCGGUAGCGUCUGCAAAG-UA----------UUU--CAAUAUCGGAUGAGUCAGAAAAAUGU ((((((..(((((((....(((((((........))).))))......)))))))...))))))......-..----------(((--(....((....))....))))..... ( -27.80) >DroSec_CAF1 747 101 - 1 GGACGUGAUGCCGGGAAAGUCGGUGAACAUGGUGUCACCCGAAAUGUACUCGGCGGUAGCGUCUGCAAAG-UA----------UUU--CGAUAUCGGGUGAGUCACAAAAAUGU ....((((((((((.....)))))..........((((((((.(((((((..((((......))))..))-))----------)..--..)).)))))))))))))........ ( -33.60) >DroSim_CAF1 764 101 - 1 GGACGUGAUGCCGGGAAAGUCGGUGAACAUGGUGUCACCCGAAAUGUACUCGGCGGUAGCGUCUGCAAAG-UA----------UUU--CGAUAUCGGGUGAGUCACAAAAAUGU ....((((((((((.....)))))..........((((((((.(((((((..((((......))))..))-))----------)..--..)).)))))))))))))........ ( -33.60) >DroEre_CAF1 777 107 - 1 GGACGUGAUGCCGGGGAAGUCGGUGAACAUGGUGUCGCCCGAAAUGUACUCAGCGGUAGCGUCUGCAAAA-UACAAA--CAU--AU--UGAUUUUGGGUGAGUCAGAUAAAUGU .(((((...(((((.....)))))..))......(((((((((((.......((((......))))....-..(((.--...--.)--)))))))))))))))).......... ( -28.80) >DroYak_CAF1 794 96 - 1 GGACGUGAUGCCGGGGAAGUCGGUGAACAUGGUGUCGCCCGAAAUGUACUCAGCGGUAGCGUCUGCAAGA-UACAAGUACAUAUUU--UGAUUUCAAGU--------------- ((((((..(((.(((....(((((((........))).))))......))).)))...)))))).(((((-((........)))))--)).........--------------- ( -24.70) >DroMoj_CAF1 500 100 - 1 GGAAGUGAUGCCAGGGAAGUCAGUAAAUAUGGAGUCGCCGGAAAUGUACUCGGCAGUGGCAUCUGGAAUUGAACAA----UU-GUUAUUAACACUGUGGGAGGGG--------- ...((((((((((..((..(((.......)))..))(((((........)))))..)))))))........(((..----..-)))......)))..........--------- ( -22.20) >consensus GGACGUGAUGCCGGGAAAGUCGGUGAACAUGGUGUCACCCGAAAUGUACUCGGCGGUAGCGUCUGCAAAG_UA__________UUU__CGAUAUCGGGUGAGUCA_AAAAAUGU ((((((..(((((((....(((((((........))).))))......)))))))...)))))).................................................. (-21.77 = -22.38 + 0.61)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:02 2006