
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
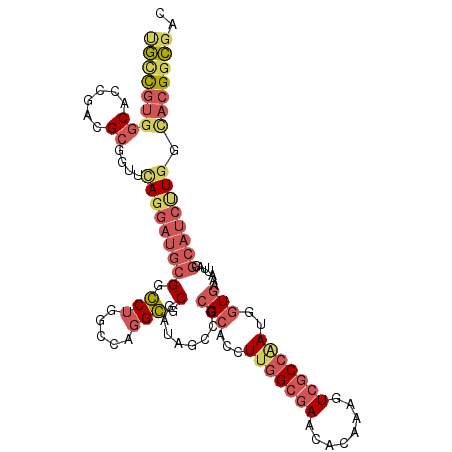
| Location | 9,776,683 – 9,776,793 |
| Length | 110 |
| Max. P | 0.787413 |

| Location | 9,776,683 – 9,776,793 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -44.20 |
| Consensus MFE | -27.76 |
| Energy contribution | -29.47 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9776683 110 + 27905053 UGCCGUGGCACCGAUGCGGUUGAGGAUGCGGCCUGGCCAGGGGCGAUAGCGGCGCAACUUGGCGAACACAAAGUCGCCAAUGGUGAAAUUAGGCAUCUUGGCUCGGCGAC .((((.(((...((((((((((......)))))(.((((..((((((...(.(((......)))....)...))))))..)))).)......)))))...)))))))... ( -44.10) >DroGri_CAF1 86103 97 + 1 CAUUUUGCCC---UCGUCUUGCAGAAUACGCGCCGGCCACGCACAAAAGCCGGGCACCUUGGCGAACACAAAGUCUCCCACUUUGAUU----------UGGUACGGUGAC .(((((((..---.......)))))))...(((((((((.........((((((...)))))).....((((((.....))))))...----------)))).))))).. ( -27.60) >DroSec_CAF1 99875 110 + 1 UGCCGUGGCACCGAUGCGUUUCAGGAUGCGGCCUGGCCAGGGGCGAUAGCCGCGCACCUUGGCAAACACAAAGUCGCCAAUGGUGAAAUUAGGCAUCUUGGCACGGCGAC .((((((.((..(((((((((((((......))))((((((((((.......))).)))))))))))......(((((...)))))......))))).)).))))))... ( -50.10) >DroSim_CAF1 100471 110 + 1 UGCCGUGGCACCGAUGCGUUUCAGGAUGCGGCCUGGCCAGGGGCGAUAGCCGCGCACCUUGGCGAACACAAAGUCGCCAAUGGUGAAAUUAAGCAUCUUGGCACGGCGAC .(((((((((....)))...((((((((((((...))).(.(((....))).).(((((((((((........))))))).)))).......)))))))))))))))... ( -52.40) >DroEre_CAF1 104164 110 + 1 UGCCGUAGCACCGACGCGGUUCAGGAUGCGGGCUGGCCAGGCGCGAUACCCGCGCACCUUAGCGAACACAAAGUCGCCGAUCGUGAAAAUAGGCAUCCUGGCACGUCGGC (((....)))((((((.(..(((((((((..(((((....(((((.....)))))...)))))..........((((.....))))......)))))))))).)))))). ( -42.70) >DroYak_CAF1 103581 110 + 1 GGCCGUGGCACCGACGCGGUUUAGGAUGCGGGCUGGCCAGGCGCGAUACCCGCGCACCUUGGCGAACACAAAGUCGCCAAUCGUGAAAAUAGGCAUCCUGGCACGGCGAC .((((((.(((((...))))..(((((((.((....))..(((((.....)))))...(((((((........)))))))............)))))))).))))))... ( -48.30) >consensus UGCCGUGGCACCGACGCGGUUCAGGAUGCGGCCUGGCCAGGCGCGAUAGCCGCGCACCUUGGCGAACACAAAGUCGCCAAUGGUGAAAUUAGGCAUCUUGGCACGGCGAC (((((((((......))....(((((((((.(((.....))).)........(((...(((((((........)))))))..))).......)))))))).))))))).. (-27.76 = -29.47 + 1.70)


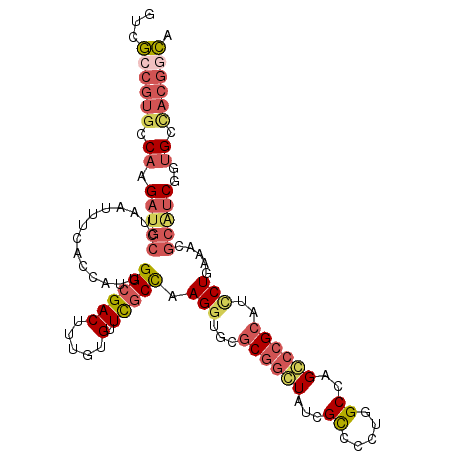
| Location | 9,776,683 – 9,776,793 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -43.82 |
| Consensus MFE | -29.17 |
| Energy contribution | -30.63 |
| Covariance contribution | 1.46 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9776683 110 - 27905053 GUCGCCGAGCCAAGAUGCCUAAUUUCACCAUUGGCGACUUUGUGUUCGCCAAGUUGCGCCGCUAUCGCCCCUGGCCAGGCCGCAUCCUCAACCGCAUCGGUGCCACGGCA ...((((.((...(((((............((((((((.....).)))))))..((((((((((.......))))..)).)))).........)))))...))..)))). ( -36.10) >DroGri_CAF1 86103 97 - 1 GUCACCGUACCA----------AAUCAAAGUGGGAGACUUUGUGUUCGCCAAGGUGCCCGGCUUUUGUGCGUGGCCGGCGCGUAUUCUGCAAGACGA---GGGCAAAAUG (((..(((....----------........((((..((((((.(....))))))).))))((....(((((((.....)))))))...))...))).---.)))...... ( -28.30) >DroSec_CAF1 99875 110 - 1 GUCGCCGUGCCAAGAUGCCUAAUUUCACCAUUGGCGACUUUGUGUUUGCCAAGGUGCGCGGCUAUCGCCCCUGGCCAGGCCGCAUCCUGAAACGCAUCGGUGCCACGGCA ...((((((.((.(((((.......((((.((((((((.....).))))))))))).((((((...(((...)))..))))))..........)))))..)).)))))). ( -49.60) >DroSim_CAF1 100471 110 - 1 GUCGCCGUGCCAAGAUGCUUAAUUUCACCAUUGGCGACUUUGUGUUCGCCAAGGUGCGCGGCUAUCGCCCCUGGCCAGGCCGCAUCCUGAAACGCAUCGGUGCCACGGCA ...((((((.((.(((((.......((((.((((((((.....).))))))))))).((((((...(((...)))..))))))..........)))))..)).)))))). ( -52.10) >DroEre_CAF1 104164 110 - 1 GCCGACGUGCCAGGAUGCCUAUUUUCACGAUCGGCGACUUUGUGUUCGCUAAGGUGCGCGGGUAUCGCGCCUGGCCAGCCCGCAUCCUGAACCGCGUCGGUGCUACGGCA (((((((((.((((((((.......(((((..(....).)))))...((((.((((((.......))))))))))......))))))))...)))))))))((....)). ( -50.40) >DroYak_CAF1 103581 110 - 1 GUCGCCGUGCCAGGAUGCCUAUUUUCACGAUUGGCGACUUUGUGUUCGCCAAGGUGCGCGGGUAUCGCGCCUGGCCAGCCCGCAUCCUAAACCGCGUCGGUGCCACGGCC ...((((((.((((((((............((((((((.....).)))))))((((((((.....)))))...))).....)))))))..((((...))))).)))))). ( -46.40) >consensus GUCGCCGUGCCAAGAUGCCUAAUUUCACCAUUGGCGACUUUGUGUUCGCCAAGGUGCGCGGCUAUCGCCCCUGGCCAGCCCGCAUCCUGAAACGCAUCGGUGCCACGGCA ...((((((.((.(((((..............((((((.....).))))).(((...((((((...((.....))..))))))..))).....)))))..)).)))))). (-29.17 = -30.63 + 1.46)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:00 2006