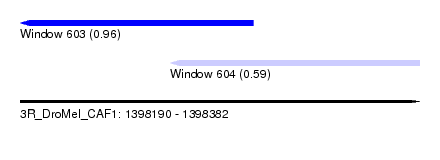
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,398,190 – 1,398,382 |
| Length | 192 |
| Max. P | 0.958958 |

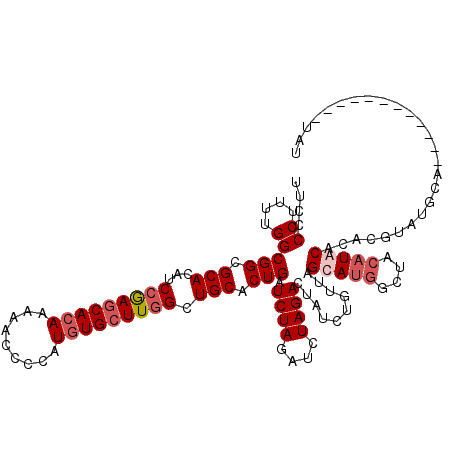
| Location | 1,398,190 – 1,398,302 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.04 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -12.79 |
| Energy contribution | -14.10 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.958958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1398190 112 - 27905053 GCAUGGCUACAUGCAUACGUAUGUAUGUACACACAUAUAUGUAAAUACGCGGAAGGAGUGUGUUUAAACAUACAUAAUGCACACAAGUGGGCUCGCAGGUA--------CAUAAUGCAAU (((((....)))))....((((.(((((((..(((....)))......((((.....((((((....)))))).....((.((....)).))))))..)))--------))))))))... ( -30.50) >DroSec_CAF1 3024 97 - 1 GCAUGGCUACAUACACACGUAUGUA------------UAUGUAAAUACGCGGAACGAGUGUGUUUAAACAUACACA-UACAUUCA--UGGGCUCGUAGGUA--------CAUAAUGCAAU ((((...(((((((....)))))))------------((((((..((((.((..(((((((((............)-))))))).--.)..))))))..))--------))))))))... ( -28.10) >DroSim_CAF1 3084 97 - 1 GCAUGGCUACAUACACACGUAUGCA------------UAUGUAAAUUCGCGGAACGAGUGUGUUUAAACAUACACA-UACAUACA--AUGGCUCGUAGGUA--------CAUAAUGCAAU ((((((((((((((....)))))..------------((((((...(((.....)))((((((....))))))...-))))))..--.))))).((....)--------)...))))... ( -21.50) >DroYak_CAF1 3023 102 - 1 GCAUGGGUA--UGCAUACAUAUACA------------UAUGUAAAAACGCGGACCGAGUGUGUUUUUAUAUA----UUGCACACAAGUGCGCUCGUAGAUAUGUACAUAUAUAAUGCAAU ((((..(((--((..(((((((..(------------((((((((((((((.......))))))))))))))----).((((....))))........))))))))))))...))))... ( -32.40) >consensus GCAUGGCUACAUACACACGUAUGCA____________UAUGUAAAUACGCGGAACGAGUGUGUUUAAACAUACACA_UACACACA__UGGGCUCGUAGGUA________CAUAAUGCAAU ((((...(((((((....)))))))............(((((((((((((.......))))))))..))))).........................................))))... (-12.79 = -14.10 + 1.31)

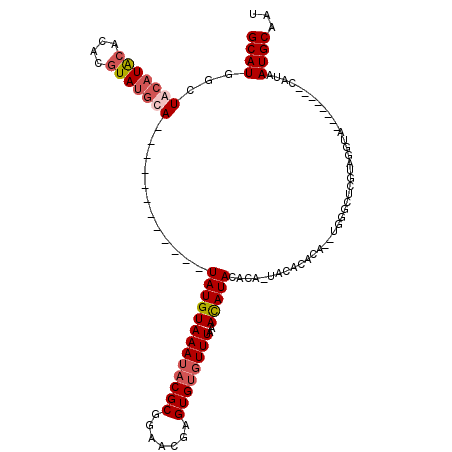
| Location | 1,398,262 – 1,398,382 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.26 |
| Mean single sequence MFE | -27.41 |
| Consensus MFE | -19.16 |
| Energy contribution | -20.73 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1398262 120 - 27905053 UUCCCCUUUUGGCGGCGCACAUCCGAGCACAAAAACCCCAUCUGCUUGGCUGCACUGAUCUAGAUCUAGACUAUCUUUUAGCAUGGCUACAUGCAUACGUAUGUAUGUACACACAUAUAU ....((....))(((.(((...(((((((.............))))))).))).))).((((....))))..........(((((....)))))......((((((((....)))))))) ( -28.42) >DroSec_CAF1 3093 108 - 1 UUCCCCUUUUGGCGGCGCACAUCCGAGCACAAAAACCCCAUGUGCUUGGCUGCACUGAUCUAGAUCUAGACUAUCUGUUAGCAUGGCUACAUACACACGUAUGUA------------UAU ....((..(((((((.(((...(((((((((.........))))))))).))).....((((....))))....)))))))...)).(((((((....)))))))------------... ( -29.60) >DroSim_CAF1 3153 108 - 1 UUCCCCUUUUGGCGGCGCACAUCCGAGCACAAAAACCCCAUGUGCUUGGCUGCACUGAUCUAGAUCUAGACUAUCUGUUAGCAUGGCUACAUACACACGUAUGCA------------UAU .........((((.(((((...(((((((((.........))))))))).))).....((((....))))..........))...))))(((((....)))))..------------... ( -26.90) >DroYak_CAF1 3099 105 - 1 UUCCCCU-UUGGCGGCGCACGUCGAAACACAAAAACCUUAUGUGCUUGGAUGCACUGAUCUAGAUCUAGACUAUCUUUUAGCAUGGGUA--UGCAUACAUAUACA------------UAU ......(-((((((.....)))))))............((((((..((.(((((...(((((...(((((......)))))..))))).--))))).))..))))------------)). ( -24.70) >consensus UUCCCCUUUUGGCGGCGCACAUCCGAGCACAAAAACCCCAUGUGCUUGGCUGCACUGAUCUAGAUCUAGACUAUCUGUUAGCAUGGCUACAUACACACGUAUGCA____________UAU ....((....))(((.(((...(((((((((.........))))))))).))).))).((((....))))..........(((((....))))).......................... (-19.16 = -20.73 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:24 2006