
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,397,077 – 1,397,458 |
| Length | 381 |
| Max. P | 0.973527 |

| Location | 1,397,077 – 1,397,192 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.40 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -23.79 |
| Energy contribution | -24.72 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1397077 115 + 27905053 UGUGU----GUGUAUACUUGUGCGCAAUGCGUGGGUAUUUUCUCAGUUUUUUGUUCUAUUUUCAAAUUUUAUUGCUCUCGCAUAAUGGUAUACAUAUGUACAUCAUUAUGUUUGACCGU ..((.----(...(((((..(((.....)))..)))))...).))............................(.((..((((((((((.(((....))).))))))))))..)).).. ( -24.90) >DroSec_CAF1 1978 119 + 1 UGUGUGUAUGUGUAUACUUGUGCGCAAUGCGUGGGUAUUUUCUCAGUUUUUUGUUCUAUUUUCAAAUUUUAUUGCUCUCGCAUAAUAGUAUACAUAUGUACAGCAUUAUGUUUGACCGU ((((((((((((((((((((((((....(((((((......))))....((((.........))))......)))...))))))..)))))))))))))))).)).............. ( -28.00) >DroSim_CAF1 1977 119 + 1 UGUGUGUAUGUGUAUACUUGUGCGCAAUGCGUGGGUAUUUUCUCAGUUUUUCGUUCUAUUUUCAAAUUUUAUUGCUCUCGCAUAAUAGUAUACAUAUGUACAGCAUUAUGUUUGACCGU ((((((((((((((((((((((((....(((((((......))))(.....)....................)))...))))))..)))))))))))))))).)).............. ( -26.50) >DroYak_CAF1 1935 117 + 1 UGUGUG--UGUGUAUACUUGUGCGCUAUGCGUGGGUAUUUUCACAGUUUUUCGUUCUAUUUUAAAAUUUUAUUGCUAUCGCACAAUAGUAUACACAUGUACAACACUAUGUUUGACCGU ((((..--((((((((((((((((.((.(((((((....)))))((((((............)))))).....)))).))))))..))))))))))..))))................. ( -34.20) >consensus UGUGUG__UGUGUAUACUUGUGCGCAAUGCGUGGGUAUUUUCUCAGUUUUUCGUUCUAUUUUCAAAUUUUAUUGCUCUCGCAUAAUAGUAUACAUAUGUACAGCAUUAUGUUUGACCGU ((((((((((((((((((((((((....(((((((......))))(.....)....................)))...))))))..)))))))))))))))).)).............. (-23.79 = -24.72 + 0.94)

| Location | 1,397,077 – 1,397,192 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.40 |
| Mean single sequence MFE | -19.18 |
| Consensus MFE | -15.06 |
| Energy contribution | -14.50 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1397077 115 - 27905053 ACGGUCAAACAUAAUGAUGUACAUAUGUAUACCAUUAUGCGAGAGCAAUAAAAUUUGAAAAUAGAACAAAAAACUGAGAAAAUACCCACGCAUUGCGCACAAGUAUACAC----ACACA .((((.....((((((.(((((....))))).))))))((....))..........................)))).....((((....((.....))....))))....----..... ( -16.40) >DroSec_CAF1 1978 119 - 1 ACGGUCAAACAUAAUGCUGUACAUAUGUAUACUAUUAUGCGAGAGCAAUAAAAUUUGAAAAUAGAACAAAAAACUGAGAAAAUACCCACGCAUUGCGCACAAGUAUACACAUACACACA ((((.((.......)))))).....((((((((....(((....)))..........................................((.....))...)))))))).......... ( -16.50) >DroSim_CAF1 1977 119 - 1 ACGGUCAAACAUAAUGCUGUACAUAUGUAUACUAUUAUGCGAGAGCAAUAAAAUUUGAAAAUAGAACGAAAAACUGAGAAAAUACCCACGCAUUGCGCACAAGUAUACACAUACACACA ((((.((.......)))))).....((((((((....(((....)))..........................................((.....))...)))))))).......... ( -16.50) >DroYak_CAF1 1935 117 - 1 ACGGUCAAACAUAGUGUUGUACAUGUGUAUACUAUUGUGCGAUAGCAAUAAAAUUUUAAAAUAGAACGAAAAACUGUGAAAAUACCCACGCAUAGCGCACAAGUAUACACA--CACACA .............((((.((....(((((((((..((((((.((((......(((((...((((.........)))).)))))......)).)).))))))))))))))))--))))). ( -27.30) >consensus ACGGUCAAACAUAAUGCUGUACAUAUGUAUACUAUUAUGCGAGAGCAAUAAAAUUUGAAAAUAGAACAAAAAACUGAGAAAAUACCCACGCAUUGCGCACAAGUAUACACA__CACACA .((((.....((((((.(((((....))))).))))))((....))..........................)))).....((((....((.....))....))))............. (-15.06 = -14.50 + -0.56)

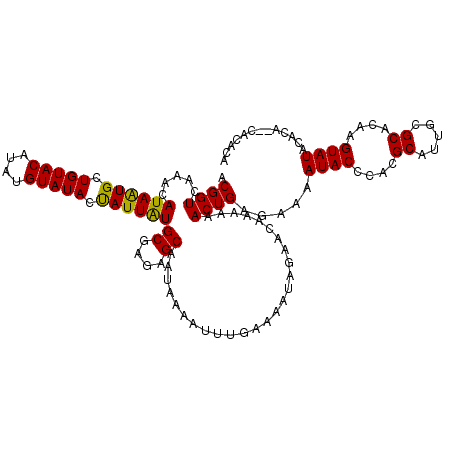
| Location | 1,397,112 – 1,397,223 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.55 |
| Mean single sequence MFE | -20.68 |
| Consensus MFE | -14.84 |
| Energy contribution | -14.40 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1397112 111 - 27905053 AAAAAACUCGCGGUGUU-------GGAG--AUUCGAAAUAACGGUCAAACAUAAUGAUGUACAUAUGUAUACCAUUAUGCGAGAGCAAUAAAAUUUGAAAAUAGAACAAAAAACUGAGAA .......((.((((.((-------(.(.--.((((((.............((((((.(((((....))))).))))))((....)).......))))))..)....)))...)))).)). ( -21.00) >DroSec_CAF1 2017 113 - 1 AAAAAACUCGCGGUGUU-------GGAGAAAUUCGAAAUAACGGUCAAACAUAAUGCUGUACAUAUGUAUACUAUUAUGCGAGAGCAAUAAAAUUUGAAAAUAGAACAAAAAACUGAGAA ......(((.((...((-------(((....))))).....)).(((((.((((((.(((((....))))).))))))((....)).......))))).................))).. ( -20.80) >DroSim_CAF1 2016 113 - 1 AAAAAACUCGCGGUGUU-------GGAGAAAUUCGAAAUAACGGUCAAACAUAAUGCUGUACAUAUGUAUACUAUUAUGCGAGAGCAAUAAAAUUUGAAAAUAGAACGAAAAACUGAGAA ......(((.((...((-------(((....))))).....)).(((((.((((((.(((((....))))).))))))((....)).......))))).................))).. ( -20.80) >DroYak_CAF1 1972 118 - 1 AA--AACUCGCGAUUGUCAUGUGCAGCGAAAUUUCAAUUAACGGUCAAACAUAGUGUUGUACAUGUGUAUACUAUUGUGCGAUAGCAAUAAAAUUUUAAAAUAGAACGAAAAACUGUGAA ..--...(((((.....((((((((((((....)).......(......).....))))))))))......(((((.(((....)))............)))))..........))))). ( -20.12) >consensus AAAAAACUCGCGGUGUU_______GGAGAAAUUCGAAAUAACGGUCAAACAUAAUGCUGUACAUAUGUAUACUAUUAUGCGAGAGCAAUAAAAUUUGAAAAUAGAACAAAAAACUGAGAA ......(((((.............(((....)))................((((((.(((((....))))).)))))))))))..................................... (-14.84 = -14.40 + -0.44)

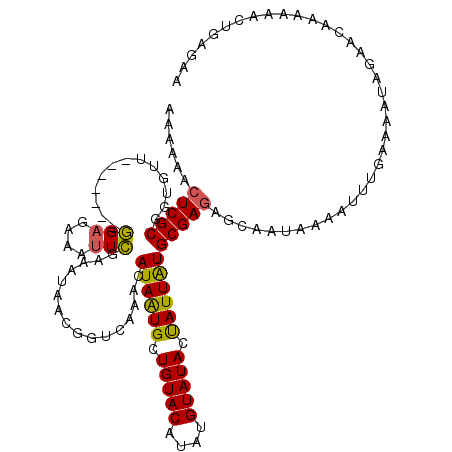
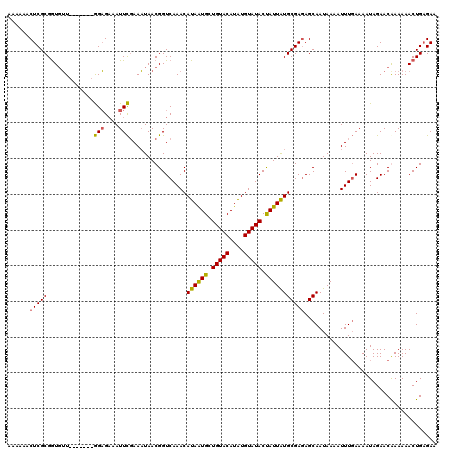
| Location | 1,397,223 – 1,397,343 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.81 |
| Mean single sequence MFE | -32.25 |
| Consensus MFE | -26.01 |
| Energy contribution | -26.20 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1397223 120 - 27905053 GUGCAUUGCACCUGAUUAGCGGGUUGUAAGUGCAGUUGUACAAACAUGCCUAUCCACAUACAUAUGUACAUACAAAUGUAUAGAGAUUGCUUACAAAUCGUGAGUGGGAAAUGCCGUCAC (.(((((...((((....(((..((((((((((((((.....))).)))..(((...((((((.(((....))).))))))...))).))))))))..)))...)))).))))))..... ( -31.80) >DroSec_CAF1 2130 116 - 1 GUGCAUUGCACCUGAUUAGCGGGUUGUAAGUGCAGUUGUGCAAACAUGCCUAUCCACA----UAUGUACAUACAAAUGUACAGAGAUGGCUUACAAAUCGGGACUGGGAAAUGCCGUCAC (.(((((...(((..((..((..((((((((((((((.....))).))).......((----(.(((((((....)))))))...)))))))))))..)).))..))).))))))..... ( -33.50) >DroSim_CAF1 2129 116 - 1 GUGCAUUGCACCUGAUUAGCGGGUUGUAAGUGCAGUUGUACAAACAUGCCUAUCCACA----UAUGUACAUACAAAUGUACAGAGAUGGCUUACAAAUCGGGACUGGGAAAUGCCGUCAC (.(((((...(((..((..((..((((((((((((((.....))).))).......((----(.(((((((....)))))))...)))))))))))..)).))..))).))))))..... ( -33.50) >DroYak_CAF1 2090 116 - 1 GUACAUUGCACCUGAUUAGCGGGUUGUAAGUGCAGUUGUACAAACAUACCUAUCCACA----UAUGUACACUUGUAUGUACAUAAAUUGCUUACAAAUCGGGACUGGUAAAUGCCGUCAC ..........(((((((((((((((((..((((....))))..))).)))).......----((((((((......))))))))....)))....)))))))...(((....)))..... ( -30.20) >consensus GUGCAUUGCACCUGAUUAGCGGGUUGUAAGUGCAGUUGUACAAACAUGCCUAUCCACA____UAUGUACAUACAAAUGUACAGAGAUGGCUUACAAAUCGGGACUGGGAAAUGCCGUCAC ..(((((.(((((((((((((((((((..((((....))))..))).)))).............(((((((....)))))))......)))....)))))))..))...)))))...... (-26.01 = -26.20 + 0.19)

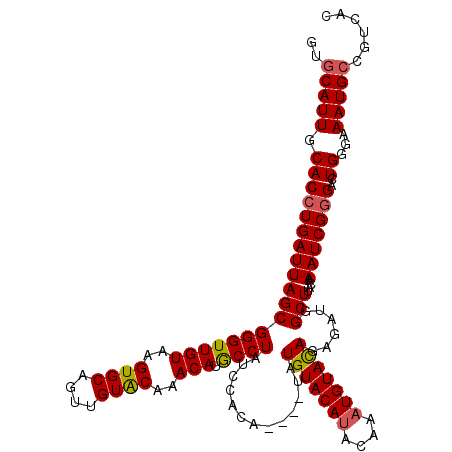
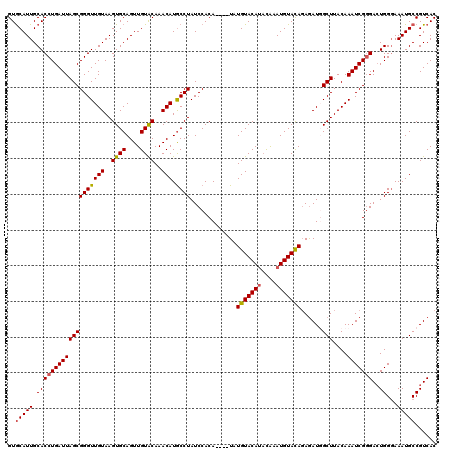
| Location | 1,397,263 – 1,397,379 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.09 |
| Mean single sequence MFE | -28.75 |
| Consensus MFE | -23.19 |
| Energy contribution | -22.25 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1397263 116 + 27905053 UACAUUUGUAUGUACAUAUGUAUGUGGAUAGGCAUGUUUGUACAACUGCACUUACAACCCGCUAAUCAGGUGCAAUGCACAUA----CAUUCAUCGAUUCCGACUCGGAAUCCCAAUCGG ......(((((((.(((.(((((.(((.((((..(((..((.(....).))..)))...).))).))).)))))))).)))))----))......(((((((...)))))))........ ( -28.90) >DroSec_CAF1 2170 112 + 1 UACAUUUGUAUGUACAUA----UGUGGAUAGGCAUGUUUGCACAACUGCACUUACAACCCGCUAAUCAGGUGCAAUGCACAUA----CAUCCAUCGAUUCCGAUUCGGAAUCCCAAUCGG ......(((((((.(((.----((..((((....))))..))....(((((((..............)))))))))).)))))----))......(((((((...)))))))........ ( -29.04) >DroSim_CAF1 2169 112 + 1 UACAUUUGUAUGUACAUA----UGUGGAUAGGCAUGUUUGUACAACUGCACUUACAACCCGCUAAUCAGGUGCAAUGCACAUA----CAUCCAUCGAUUCCGAUUCGGAAUCCCAAUCGG (((((.(((....))).)----))))(((.((.((((.(((.((..(((((((..............))))))).)).))).)----))))))))(((((((...)))))))........ ( -29.14) >DroYak_CAF1 2130 112 + 1 UACAUACAAGUGUACAUA----UGUGGAUAGGUAUGUUUGUACAACUGCACUUACAACCCGCUAAUCAGGUGCAAUGUACAUAUAAACAUCCAUCGAUUCCGAUUCGGAAUUCCAA---- (((((....)))))....----..((((..((.(((((((((((..(((((((..............))))))).))))))....)))))))....((((((...)))))))))).---- ( -27.94) >consensus UACAUUUGUAUGUACAUA____UGUGGAUAGGCAUGUUUGUACAACUGCACUUACAACCCGCUAAUCAGGUGCAAUGCACAUA____CAUCCAUCGAUUCCGAUUCGGAAUCCCAAUCGG (((((....))))).........((((((.((..(((.((((....))))...)))..)).........((((...))))........)))))).(((((((...)))))))........ (-23.19 = -22.25 + -0.94)

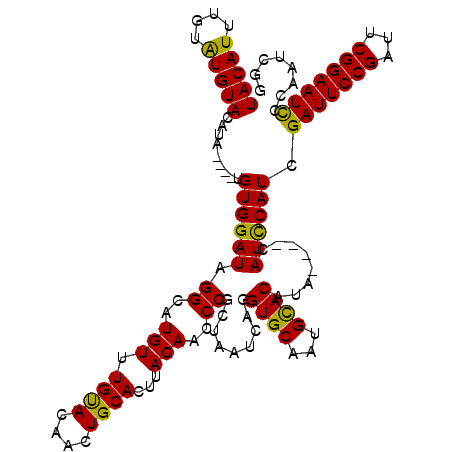
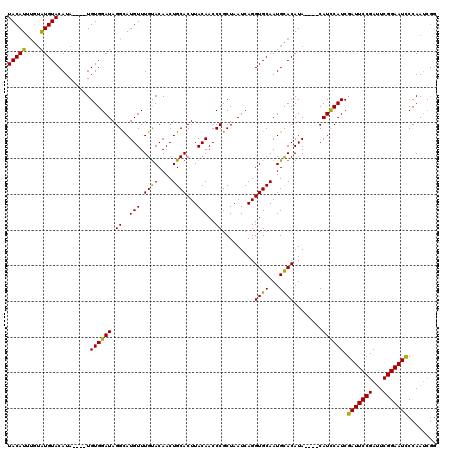
| Location | 1,397,263 – 1,397,379 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.09 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -27.33 |
| Energy contribution | -27.70 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1397263 116 - 27905053 CCGAUUGGGAUUCCGAGUCGGAAUCGAUGAAUG----UAUGUGCAUUGCACCUGAUUAGCGGGUUGUAAGUGCAGUUGUACAAACAUGCCUAUCCACAUACAUAUGUACAUACAAAUGUA ((((((.((...)).))))))..........((----(((((((((((((((((.....)))).))))((.((((((.....))).)))))............)))))))))))...... ( -35.10) >DroSec_CAF1 2170 112 - 1 CCGAUUGGGAUUCCGAAUCGGAAUCGAUGGAUG----UAUGUGCAUUGCACCUGAUUAGCGGGUUGUAAGUGCAGUUGUGCAAACAUGCCUAUCCACA----UAUGUACAUACAAAUGUA ((((((.((...)).))))))..........((----(((((((((((((((((.....)))).))))((.((((((.....))).))))).......----.)))))))))))...... ( -35.10) >DroSim_CAF1 2169 112 - 1 CCGAUUGGGAUUCCGAAUCGGAAUCGAUGGAUG----UAUGUGCAUUGCACCUGAUUAGCGGGUUGUAAGUGCAGUUGUACAAACAUGCCUAUCCACA----UAUGUACAUACAAAUGUA ((((((.((...)).))))))..........((----(((((((((((((((((.....)))).))))((.((((((.....))).))))).......----.)))))))))))...... ( -35.10) >DroYak_CAF1 2130 112 - 1 ----UUGGAAUUCCGAAUCGGAAUCGAUGGAUGUUUAUAUGUACAUUGCACCUGAUUAGCGGGUUGUAAGUGCAGUUGUACAAACAUACCUAUCCACA----UAUGUACACUUGUAUGUA ----..(((((((((...))))))..(((..(((.....(((((.(((((((((.....)))).)))))))))).....)))..))).....)))(((----((((......))))))). ( -31.10) >consensus CCGAUUGGGAUUCCGAAUCGGAAUCGAUGGAUG____UAUGUGCAUUGCACCUGAUUAGCGGGUUGUAAGUGCAGUUGUACAAACAUGCCUAUCCACA____UAUGUACAUACAAAUGUA ........(((((((...)))))))(.((((((....(((((...(((((((((.....)))).)))))((((....))))..)))))..))))))).........(((((....))))) (-27.33 = -27.70 + 0.37)

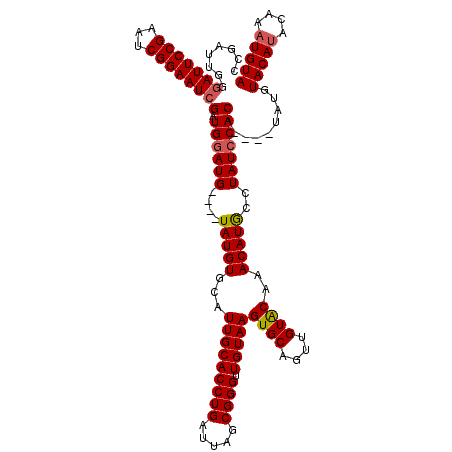
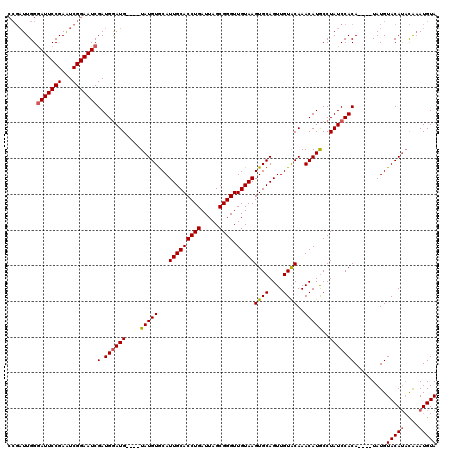
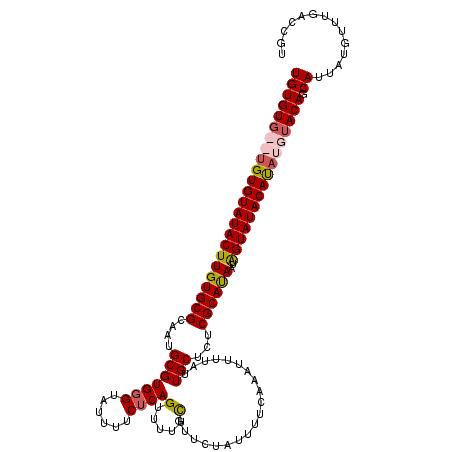
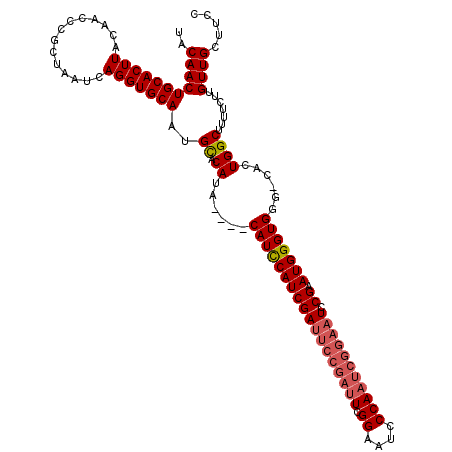
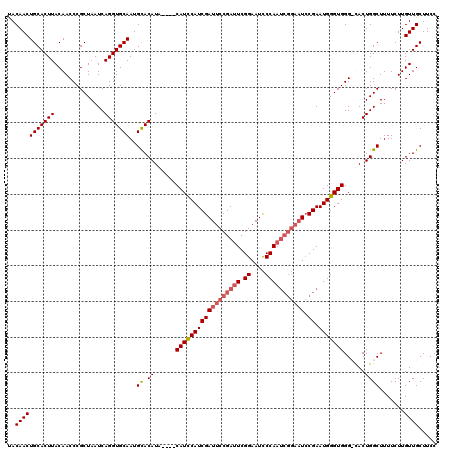
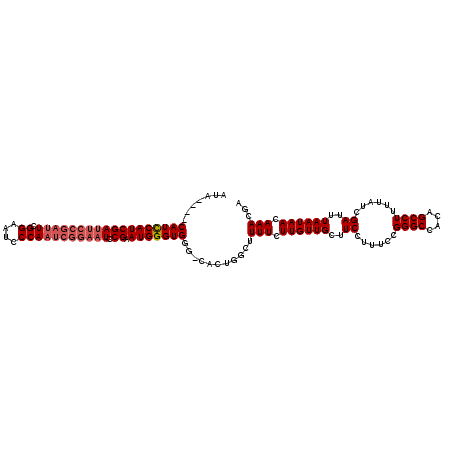
| Location | 1,397,303 – 1,397,418 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.08 |
| Mean single sequence MFE | -33.24 |
| Consensus MFE | -28.72 |
| Energy contribution | -30.09 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1397303 115 + 27905053 UACAACUGCACUUACAACCCGCUAAUCAGGUGCAAUGCACAUA----CAUUCAUCGAUUCCGACUCGGAAUCCCAAUCGGAAUCCGAAUGGGUGGG-UACUGGCUUUUCUUGUUGCUUCC ..((((........(((((((((......((((...))))...----(((((...((((((((...((....))..)))))))).)))))))))))-)..)).........))))..... ( -33.83) >DroSec_CAF1 2206 116 + 1 CACAACUGCACUUACAACCCGCUAAUCAGGUGCAAUGCACAUA----CAUCCAUCGAUUCCGAUUCGGAAUCCCAAUCGGAAUCCGAAUGGGUGGGCCACUGGCUUUUCUUGUUGCUUCC ......(((((((..............)))))))..(((((..----((((((((((((((((((.((....))))))))))).)).)))))))((((...)))).....)).))).... ( -37.24) >DroSim_CAF1 2205 116 + 1 UACAACUGCACUUACAACCCGCUAAUCAGGUGCAAUGCACAUA----CAUCCAUCGAUUCCGAUUCGGAAUCCCAAUCGGAAUCCGAAUGGGUGGGCCACUGGCUUUUCUUGUUGCUUCC ......(((((((..............)))))))..(((((..----((((((((((((((((((.((....))))))))))).)).)))))))((((...)))).....)).))).... ( -37.24) >DroYak_CAF1 2166 113 + 1 UACAACUGCACUUACAACCCGCUAAUCAGGUGCAAUGUACAUAUAAACAUCCAUCGAUUCCGAUUCGGAAUUCCAA------UACGAAUGGGUGGG-UACUGGCUUUUCUUGUUGCUUCC ..(((((((((((..............)))))))..((((.......(((((((((((((((...)))))).....------..)).))))))).)-)))...........))))..... ( -24.65) >consensus UACAACUGCACUUACAACCCGCUAAUCAGGUGCAAUGCACAUA____CAUCCAUCGAUUCCGAUUCGGAAUCCCAAUCGGAAUCCGAAUGGGUGGG_CACUGGCUUUUCUUGUUGCUUCC ..(((((((((((..............)))))))..((.((......((((((((((((((((((.((....))))))))))).)).)))))))......)))).......))))..... (-28.72 = -30.09 + 1.37)

| Location | 1,397,303 – 1,397,418 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.08 |
| Mean single sequence MFE | -38.90 |
| Consensus MFE | -33.10 |
| Energy contribution | -34.48 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1397303 115 - 27905053 GGAAGCAACAAGAAAAGCCAGUA-CCCACCCAUUCGGAUUCCGAUUGGGAUUCCGAGUCGGAAUCGAUGAAUG----UAUGUGCAUUGCACCUGAUUAGCGGGUUGUAAGUGCAGUUGUA ....(((((..............-..((..((((((((((((((((.((...)).))))))))))..))))))----..))(((((((((((((.....)))).))).))))))))))). ( -40.80) >DroSec_CAF1 2206 116 - 1 GGAAGCAACAAGAAAAGCCAGUGGCCCACCCAUUCGGAUUCCGAUUGGGAUUCCGAAUCGGAAUCGAUGGAUG----UAUGUGCAUUGCACCUGAUUAGCGGGUUGUAAGUGCAGUUGUG ....(((((.......(((...))).((.(((((..((((((((((.((...)).))))))))))))))).))----..((..(.(((((((((.....)))).))))))..))))))). ( -42.30) >DroSim_CAF1 2205 116 - 1 GGAAGCAACAAGAAAAGCCAGUGGCCCACCCAUUCGGAUUCCGAUUGGGAUUCCGAAUCGGAAUCGAUGGAUG----UAUGUGCAUUGCACCUGAUUAGCGGGUUGUAAGUGCAGUUGUA ....(((((.......(((...))).((.(((((..((((((((((.((...)).))))))))))))))).))----..((..(.(((((((((.....)))).))))))..))))))). ( -42.30) >DroYak_CAF1 2166 113 - 1 GGAAGCAACAAGAAAAGCCAGUA-CCCACCCAUUCGUA------UUGGAAUUCCGAAUCGGAAUCGAUGGAUGUUUAUAUGUACAUUGCACCUGAUUAGCGGGUUGUAAGUGCAGUUGUA ....(((((..((....((((((-(..........)))------))))..(((((...)))))))..............(((((.(((((((((.....)))).))))))))))))))). ( -30.20) >consensus GGAAGCAACAAGAAAAGCCAGUA_CCCACCCAUUCGGAUUCCGAUUGGGAUUCCGAAUCGGAAUCGAUGGAUG____UAUGUGCAUUGCACCUGAUUAGCGGGUUGUAAGUGCAGUUGUA ....(((((.................((.((((.((.(((((((((.(.....).))))))))))))))).))......(((((.(((((((((.....)))).))))))))))))))). (-33.10 = -34.48 + 1.38)

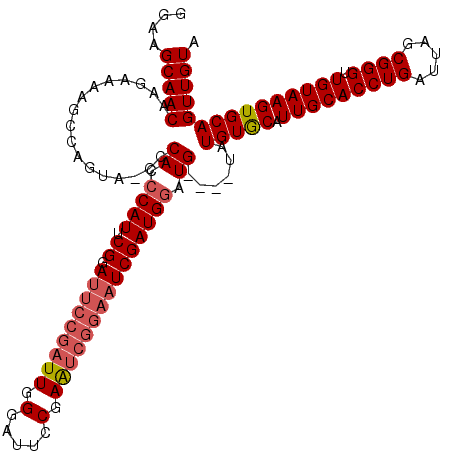
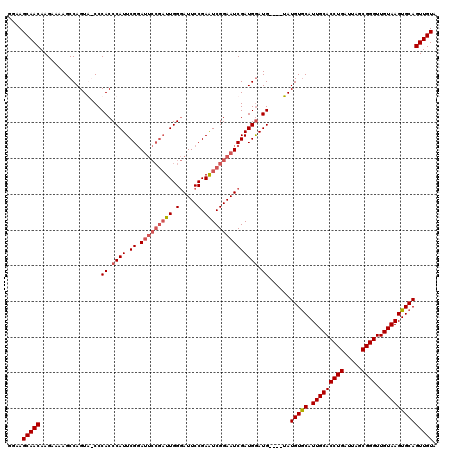
| Location | 1,397,343 – 1,397,458 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -30.33 |
| Consensus MFE | -25.81 |
| Energy contribution | -27.38 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1397343 115 + 27905053 AUA----CAUUCAUCGAUUCCGACUCGGAAUCCCAAUCGGAAUCCGAAUGGGUGGG-UACUGGCUUUUCUUGUUGCUUCCUUUACGGGCCACAGCCUUUUAUCGAUUUAAUAACAAACGA ...----((((((((((((((((...((....))..))))))).)).)))))))((-((..((((...(((((..........)))))....))))...))))................. ( -28.30) >DroSec_CAF1 2246 116 + 1 AUA----CAUCCAUCGAUUCCGAUUCGGAAUCCCAAUCGGAAUCCGAAUGGGUGGGCCACUGGCUUUUCUUGUUGCUUCCUUUCCGGGCCACAGCCUUUUAUCGAUUUAAUAACAAACGA ...----((((((((((((((((((.((....))))))))))).)).)))))))((((...))))(((.((((((..((......((((....))))......))..)))))).)))... ( -35.30) >DroSim_CAF1 2245 116 + 1 AUA----CAUCCAUCGAUUCCGAUUCGGAAUCCCAAUCGGAAUCCGAAUGGGUGGGCCACUGGCUUUUCUUGUUGCUUCCUUUCCGGGCCACAGCCUUUUAUCGAUUUAAUAACAAACGA ...----((((((((((((((((((.((....))))))))))).)).)))))))((((...))))(((.((((((..((......((((....))))......))..)))))).)))... ( -35.30) >DroYak_CAF1 2206 113 + 1 AUAUAAACAUCCAUCGAUUCCGAUUCGGAAUUCCAA------UACGAAUGGGUGGG-UACUGGCUUUUCUUGUUGCUUCCUUUCCGGGCCACAGCCUUUUAUCGAUUUAAUAACAAACGA .......(((((((((((((((...)))))).....------..)).)))))))((-((..((((......((.((((.......)))).))))))...))))................. ( -22.41) >consensus AUA____CAUCCAUCGAUUCCGAUUCGGAAUCCCAAUCGGAAUCCGAAUGGGUGGG_CACUGGCUUUUCUUGUUGCUUCCUUUCCGGGCCACAGCCUUUUAUCGAUUUAAUAACAAACGA .......((((((((((((((((((.((....))))))))))).)).)))))))...........(((.((((((..((......((((....))))......))..)))))).)))... (-25.81 = -27.38 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:23 2006