
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,730,760 – 9,730,890 |
| Length | 130 |
| Max. P | 0.633462 |

| Location | 9,730,760 – 9,730,872 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.99 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -18.63 |
| Energy contribution | -19.52 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
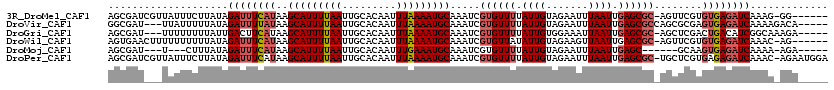
>3R_DroMel_CAF1 9730760 112 + 27905053 AGCGAUCGUUAUUUCUUAUAGAUUUCAUAAGCAUUUUAAUUGCACAAUUUAAAAUGCAAAUCGUGUUUUAUUGUAGAAUUUAAUUGAGCGC-AGUUCGUGUGAGAUCAAAG-GG------ ....................(((((((((.(((((((((.........))))))))).....((((((.((((.......)))).))))))-......)))))))))....-..------ ( -25.00) >DroVir_CAF1 41232 112 + 1 GGCGAU---UUAUUUUUAUAGAUUUUAUAAGCAUUUUAAUUGCACAAUUUAAAAUGCAAAUCGUGUUUUAUUGUAGAAUUUAAUUGAGCGCCAGCGCGAGUGAGAUCAAAAGACA----- ((((.(---(((......(((((((((((((((.......)))......((((((((.....))))))))))))))))))))..)))))))).((....))..............----- ( -23.60) >DroGri_CAF1 41290 111 + 1 AGCGAU---UUUUUUUUAUUGACUUCAUAAGCAUUUUAAUUGCACAAUUUAAAAUGCAAAUCGUGUUUUAUUGUGGAAAUUAAUUGAGCGC-AGCUCGACUGACAUCGGCAAAGA----- .(((((---((........((....))...(((((((((.........))))))))))))))))......((((.((..(((.((((((..-.)))))).)))..)).))))...----- ( -25.50) >DroWil_CAF1 38987 112 + 1 AGUGAACUUUUUUUUUUAUAGAUUUCAUAAGCAUUUUAAUUGCACAAUUUAAAAUGCAAAUCGUGUUAUAUUGUAGAAGUUAAUUGAGCGC-AGUUCGUGUGAGAUCAAAC-AG------ ....................(((((((((.(((((((((.........))))))))).....(((((..((((.......))))..)))))-......)))))))))....-..------ ( -22.70) >DroMoj_CAF1 43930 102 + 1 AGCGAU---U---CUUUAUAGAUUUCAUAAGCAUUUUAAUUGCACAAUUUGAAAUGCAAAUCGUGUUUUAUUGUAGAAUUUAAUUGAGC------GCAAGUGAGAUCAAAA-AGA----- ......---(---((((...((((((((..(((((((((.........))))))))).....((((((.((((.......)))).))))------))..))))))))..))-)))----- ( -25.20) >DroPer_CAF1 51538 118 + 1 AGCGAUCGUUAUUUCUUAUAGAUUUCAUAAGCAUUUUAAUUGCACAAUUUAAAAUGCAAAUCGUGUUUUAUUGUAGAAUUUAAUUGAGCGC-UGCUCGUGAGAGAUCAAAC-AGAAUGGA ..((.((((((((((((((.(((((.....(((((((((.........))))))))))))))((((((.((((.......)))).))))))-.....)))))))))..)))-.)).)).. ( -23.80) >consensus AGCGAU___UUUUUUUUAUAGAUUUCAUAAGCAUUUUAAUUGCACAAUUUAAAAUGCAAAUCGUGUUUUAUUGUAGAAUUUAAUUGAGCGC_AGCUCGAGUGAGAUCAAAA_AGA_____ ....................((((((((..(((((((((.........))))))))).....((((((.((((.......)))).))))))........))))))))............. (-18.63 = -19.52 + 0.89)


| Location | 9,730,800 – 9,730,890 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 72.88 |
| Mean single sequence MFE | -12.55 |
| Consensus MFE | -8.81 |
| Energy contribution | -9.17 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9730800 90 - 27905053 -UUC--AGCAUC-----CACCCCCG---C----------CC-CUUUGAUCUCACACGAACU-GCGCUCAAUUAAAUUCUACAAUAAAACACGAUUUGCAUUUUAAAUUGUGCA -...--......-----......((---(----------..-.((((........))))..-))).......................(((((((((.....))))))))).. ( -9.00) >DroVir_CAF1 41269 107 - 1 -CACCCACCCCCAAAAGCGCCCCAAAGCAGUCG-----UGUCUUUUGAUCUCACUCGCGCUGGCGCUCAAUUAAAUUCUACAAUAAAACACGAUUUGCAUUUUAAAUUGUGCA -..............((((((.....(((((.(-----.(((....))).).))).))...)))))).....................(((((((((.....))))))))).. ( -21.50) >DroPse_CAF1 51457 101 - 1 CUUC--ACUCUC-----CACUCUCC---ACUCUUCCAUUCU-GUUUGAUCUCUCACGAGCA-GCGCUCAAUUAAAUUCUACAAUAAAACACGAUUUGCAUUUUAAAUUGUGCA ....--......-----........---...........((-(((((........))))))-).........................(((((((((.....))))))))).. ( -12.40) >DroGri_CAF1 41327 90 - 1 -UAC-------------CACCCCCA---AACCG-----UCUUUGCCGAUGUCAGUCGAGCU-GCGCUCAAUUAAUUUCCACAAUAAAACACGAUUUGCAUUUUAAAUUGUGCA -...-------------........---...((-----(....((((((....)))).)).-))).......................(((((((((.....))))))))).. ( -13.00) >DroWil_CAF1 39027 89 - 1 -UAU--CG-AUA-----CGCCUCCA---U----------CU-GUUUGAUCUCACACGAACU-GCGCUCAAUUAACUUCUACAAUAUAACACGAUUUGCAUUUUAAAUUGUGCA -...--.(-(..-----(((.....---.----------..-(((((........))))).-))).))....................(((((((((.....))))))))).. ( -9.90) >DroAna_CAF1 51016 83 - 1 -----------C-----CACUCCCU---C-------A--CA-CUUUGAUCUCUCACGAGUU-GCGCUCAAUUAAAUUCUACAAUAAAACACGAUUUGCAUUUUAAAUUGUGCA -----------.-----........---.-------.--..-..............(((..-...)))....................(((((((((.....))))))))).. ( -9.50) >consensus _UAC__A___UC_____CACCCCCA___A__________CU_CUUUGAUCUCACACGAGCU_GCGCUCAAUUAAAUUCUACAAUAAAACACGAUUUGCAUUUUAAAUUGUGCA ........................................................((((....))))....................(((((((((.....))))))))).. ( -8.81 = -9.17 + 0.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:41 2006