
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,718,014 – 9,718,169 |
| Length | 155 |
| Max. P | 0.861465 |

| Location | 9,718,014 – 9,718,129 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.81 |
| Mean single sequence MFE | -37.05 |
| Consensus MFE | -31.51 |
| Energy contribution | -31.93 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9718014 115 - 27905053 GAUGCCCAUGCCAAUGGGUUGUCAUCUGGGCCAUAUAUCUUGUUAUGGCCAC-----GGCGCAUGCGCGCCUCGUCAAUUAGUUUGGCUGUCAAAAAGAUGAUGAACUCGUUGCUUGAUG ......((.((.(((((((((((((((.(((((((........)))))))..-----(((((....)))))..(((((.....)))))........))))))).)))))))))).))... ( -44.30) >DroVir_CAF1 28995 118 - 1 -CGGACCAAA-CAUUGGGUUGUCAUCUUGGCCAUAUAUCUUGUUAUGGUCGCGGCGCGGCGCAUGCGCGUCUCGUCAAUUAGUUUGGCUGUCAAAAAGAUGAUGAACUCGUUGCUUGAUG -.....(((.-((.(((((((((((((((((((((........)))))))..((((.(((((....))))).)(((((.....))))).)))...)))))))).)))))).)).)))... ( -41.10) >DroPse_CAF1 38815 102 - 1 -------------UUGGGUUGUCAUCUCGGCCAUAUAUCUUGUUAUGGCCAC-----GGCGCAUGCGCGUCUCGUCAAUUAGUUUGGCUGUCAAAAAGAUGAUGAACUCGUUGCUUGAUG -------------..((((((((((((.(((((((........)))))))..-----(((((....)))))..(((((.....)))))........))))))).)))))........... ( -35.20) >DroGri_CAF1 28079 113 - 1 -GGUACCAGG-CAUUUGGUUGUCAUCUUGGCCAUAUAUCUUGUUAUAGCCAC-----GGCGCAUGCGCGUCUCGUCAAUUAGUUUGGCUGUCAAAAAGAUGAUGAACUCGUUGCUUGAUG -.....((((-((...(((((((((((((((.(((........))).)))..-----(((((....)))))..(((((.....))))).......)))))))).))))...))))))... ( -35.70) >DroWil_CAF1 26502 113 - 1 --UCCUCCGUCGGAUGGGUUGUCAUCUUGGCCAUAUAUCUUGUUAUAGCCAC-----GCCGCAUAAGCGUCUCGUCAAUUAGUUUGGCUGUCAAAAAGAUGAUGAACUCGUUGCUUGAUG --.....((((((((((((((((((((((((.(((........))).)))((-----((.......))))...(((((.....))))).......)))))))).))))))))...))))) ( -30.80) >DroPer_CAF1 38890 102 - 1 -------------UUGGGUUGUCAUCUCGGCCAUAUAUCUUGUUAUGGCCAC-----GGCGCAUGCGCGUCUCGUCAAUUAGUUUGGCUGUCAAAAAGAUGAUGAACUCGUUGCUUGAUG -------------..((((((((((((.(((((((........)))))))..-----(((((....)))))..(((((.....)))))........))))))).)))))........... ( -35.20) >consensus _____CCA___CAUUGGGUUGUCAUCUUGGCCAUAUAUCUUGUUAUGGCCAC_____GGCGCAUGCGCGUCUCGUCAAUUAGUUUGGCUGUCAAAAAGAUGAUGAACUCGUUGCUUGAUG ..............(((((((((((((.(((((((........))))))).......(((((....)))))..(((((.....)))))........))))))).)))))).......... (-31.51 = -31.93 + 0.42)

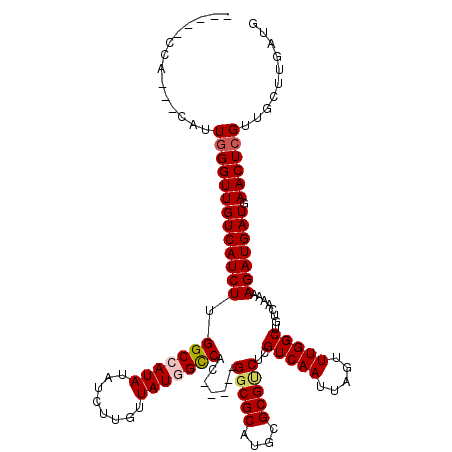

| Location | 9,718,054 – 9,718,169 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.67 |
| Mean single sequence MFE | -36.53 |
| Consensus MFE | -28.97 |
| Energy contribution | -29.42 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9718054 115 - 27905053 CUGAAGUGUAUCCGAUGCCCAAUACCACAUAUAAUAUAGCGAUGCCCAUGCCAAUGGGUUGUCAUCUGGGCCAUAUAUCUUGUUAUGGCCACGGCGCAUGCGCGCCUCGUCAAUU .............((((.......................(((((((((....))))).)))).....(((((((........)))))))..(((((....))))).)))).... ( -34.10) >DroSec_CAF1 41689 110 - 1 CCGAUG-----CCGAUGCCCAAUCCCACAUACAGUAUAGCGAUGCCCAUGCCAAUGGGUUGUCAUCUGGGCCAUAUAUCUUGUUAUGGCCACGGCGCAUGCGCGCCUCGUCAAUU .(((.(-----(((((((((...........(((.((..((..((((((....))))))))..)))))(((((((........)))))))..)).)))).)).)).)))...... ( -39.40) >DroSim_CAF1 41556 110 - 1 CCGAUG-----CCGAUGCCCAAUUCCACAUACAGUAUAGCGAUGCCCAUGCCAAUGGGUUGUCAUCUGGGCCAUAUAUCUUGUUAUGGCCACGGCGCAUGCGCGCCUCGUCAAUU .(((.(-----(((((((((...........(((.((..((..((((((....))))))))..)))))(((((((........)))))))..)).)))).)).)).)))...... ( -39.40) >DroEre_CAF1 43427 103 - 1 CGGAUA-----CCGAUGGC------CAUGC-CUAUGCAGCGAUGCCCAUGCCGAUGGGUUGUCAUCUGGGCCAUACAUCUUGUUAUGGCCACGGCGCAUGCGCGCCCCGUCAAUU (((...-----..((((((------..(((-....))).....((((((....)))))).))))))..(((((((........)))))))..(((((....))))))))...... ( -38.80) >DroYak_CAF1 43432 109 - 1 CCGAUG-----UCCAUGUCCAUACCCAUGC-CUAUACAGCGAUGCCCAUGCCGAUGGGUUGUCAUCUGGGCCAUAUAUCUUGUUAUGGCCACGGCGCAUGCGCGCCUCGUCAAUU ..((((-----..((((........)))).-.....(((.(((((((((....))))).))))..)))(((((((........)))))))..(((((....))))).)))).... ( -36.40) >DroAna_CAF1 39097 98 - 1 CGGAGU-----CUGG-------A---ACCUGGAGUA-GGCACCGACUAUGCU-AUGGGUUGUCAUCUGGGCCAUAUAUCUUGUUAUGGCCACGGCGCAUGCGCGUCUCGUCAAUU .((.((-----(((.-------.---........))-))).))(((.(((((-(((.(((((......(((((((........)))))))))))).)))).))))...))).... ( -31.10) >consensus CCGAUG_____CCGAUGCCCAAU_CCACAUACAAUAUAGCGAUGCCCAUGCCAAUGGGUUGUCAUCUGGGCCAUAUAUCUUGUUAUGGCCACGGCGCAUGCGCGCCUCGUCAAUU .............((((.......................(((((((((....))))).)))).....(((((((........)))))))..(((((....))))).)))).... (-28.97 = -29.42 + 0.45)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:36 2006