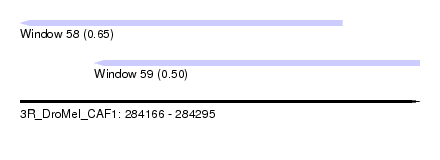
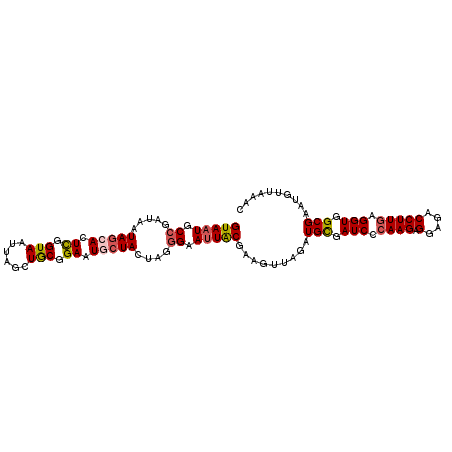
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 284,166 – 284,295 |
| Length | 129 |
| Max. P | 0.647749 |

| Location | 284,166 – 284,270 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.96 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -19.82 |
| Energy contribution | -20.30 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 284166 104 - 27905053 AUUAGCUGCGGAAUGCUACUAUGGAAUUACGAAGUUAGAUGCGAUCCCAAGAGGAGGCCUUCAGGUAGCGAAUGUUAAGCCGUUUUAUUGCCAAUAAAGCAGUA---------------- ....((((((((.(((.(((.((......)).))).....))).)))...((((....)))).(((((.(((((......)))))..)))))......))))).---------------- ( -23.90) >DroSec_CAF1 19350 104 - 1 AUUAGCUGCGGAAUGCUACUAGGGAAUUACGAAGUUAGAUGCGAUCCCAAGAGGAGACCUUGAGGUGGCGAAUGUUAAACCGUUUUAUUGCCACUAAAGCAGUA---------------- ....((((((((.(((.(((..(......)..))).....))).)))((((.(....))))).(((((((((((......)))....))))))))...))))).---------------- ( -29.50) >DroSim_CAF1 20649 104 - 1 AUUAGCUGCGGAAUGCUACUAGGGAAUUACGAAGUUAGAUGCGAUCCCAAGAGGAGACCUUGGGGUGGCGAAUGUUAAACCGUUUUAUUGCCACUAAAGCAGUA---------------- ....(((((((....))....((.(((.(((...((((.(((.((((((((.(....))))))))).)))....))))..)))...))).))......))))).---------------- ( -33.30) >DroEre_CAF1 22001 103 - 1 AUUAUCUGCGGAAUACUACUUG-GAAUUGCGAAGUUAGAUGCGAUCCCAAGAGGAGACCUUGGGGUUGCGAGUGUUAAACCAUUUUAUUGCAACUAAAGUAGUA---------------- .............((((((((.-...((((((........(((((((((((.(....))))))))))))(((((......)))))..))))))...))))))))---------------- ( -36.60) >DroYak_CAF1 21617 120 - 1 AUUAUCUACGGAAUACUACUUGGGAAUUACAAAGUUAGAUGUGAUCCCAAGAGGAAACCUUGAGGUUGCGAAUGUGAAACCGUUUUAUUGCCACUAAGGUAGUAGCUUAGCAGUGCAUUC ....((....))...((.(((((((.(((((........)))))))))))).)).((((....))))..((((((...((.(((((((((((.....))))))))...))).)))))))) ( -32.40) >consensus AUUAGCUGCGGAAUGCUACUAGGGAAUUACGAAGUUAGAUGCGAUCCCAAGAGGAGACCUUGAGGUGGCGAAUGUUAAACCGUUUUAUUGCCACUAAAGCAGUA________________ ....((((((((.(((.(((..(......)..))).....))).)))((((.(....))))).(((((((((((......)))....))))))))...)))))................. (-19.82 = -20.30 + 0.48)

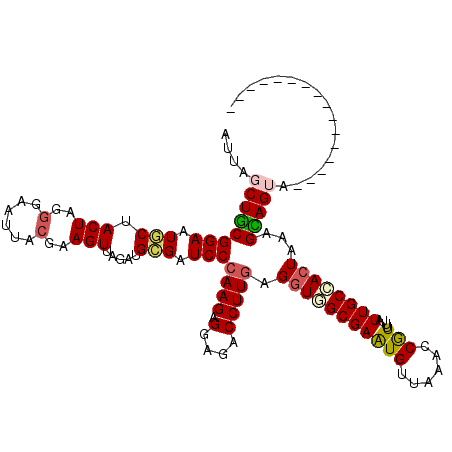
| Location | 284,190 – 284,295 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 90.29 |
| Mean single sequence MFE | -31.50 |
| Consensus MFE | -21.10 |
| Energy contribution | -21.58 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 284190 105 - 27905053 GUAAUGCCGAUAAUAGCACUCGGUAAUUAGCUGCGGAAUGCUACUAUGGAAUUACGAAGUUAGAUGCGAUCCCAAGAGGAGGCCUUCAGGUAGCGAAUGUUAAGC (((((.((.(((.(((((.((.(((......))).)).))))).))))).))))).........(((..(((.....))).(((....))).))).......... ( -24.90) >DroSec_CAF1 19374 105 - 1 GUAAUGCCGAUAAUAGCACUCGGUAAUUAGCUGCGGAAUGCUACUAGGGAAUUACGAAGUUAGAUGCGAUCCCAAGAGGAGACCUUGAGGUGGCGAAUGUUAAAC (((((.((..((.(((((.((.(((......))).)).))))).)).)).))))).........(((.(((.((((.(....))))).))).))).......... ( -30.40) >DroSim_CAF1 20673 105 - 1 GUAAUGCCGAUAAUAGCACUCGGUAAUUAGCUGCGGAAUGCUACUAGGGAAUUACGAAGUUAGAUGCGAUCCCAAGAGGAGACCUUGGGGUGGCGAAUGUUAAAC (((((.((..((.(((((.((.(((......))).)).))))).)).)).))))).........(((.((((((((.(....))))))))).))).......... ( -37.30) >DroEre_CAF1 22025 104 - 1 GUACUGCCGAUAAUAGCAGUUGGUAAUUAUCUGCGGAAUACUACUUG-GAAUUGCGAAGUUAGAUGCGAUCCCAAGAGGAGACCUUGGGGUUGCGAGUGUUAAAC ...((((.((((((.((.....)).)))))).))))((((((((((.-(.....).))))....((((((((((((.(....))))))))))))))))))).... ( -39.40) >DroYak_CAF1 21657 105 - 1 GUAAUGCCAAUAAUAACACUUGGUAAUUAUCUACGGAAUACUACUUGGGAAUUACAAAGUUAGAUGUGAUCCCAAGAGGAAACCUUGAGGUUGCGAAUGUGAAAC ................(((((.((((((.((...((....((.(((((((.(((((........))))))))))))))....))..)))))))).)).))).... ( -25.50) >consensus GUAAUGCCGAUAAUAGCACUCGGUAAUUAGCUGCGGAAUGCUACUAGGGAAUUACGAAGUUAGAUGCGAUCCCAAGAGGAGACCUUGAGGUGGCGAAUGUUAAAC (((((.((.....(((((.((.(((......))).)).)))))....)).))))).........(((.(((.((((.(....))))).))).))).......... (-21.10 = -21.58 + 0.48)

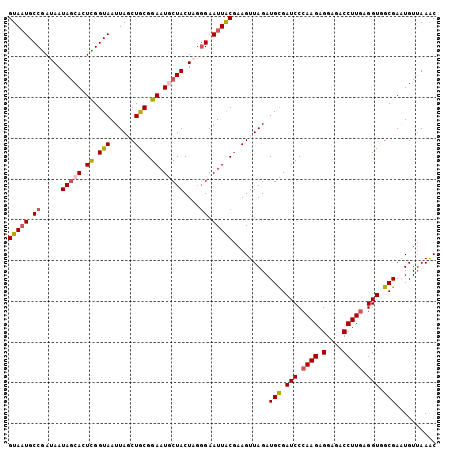
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:58 2006