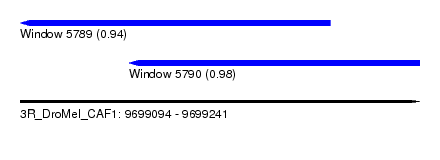
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,699,094 – 9,699,241 |
| Length | 147 |
| Max. P | 0.978689 |

| Location | 9,699,094 – 9,699,208 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.37 |
| Mean single sequence MFE | -37.09 |
| Consensus MFE | -16.92 |
| Energy contribution | -17.47 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9699094 114 - 27905053 CCACA------UCCGCAUCUGCCGCCCGGCCAAAUUCCACUUGGAAUCUUUCCCGGCGGACCGCAUCCCGGGCAUCCUCCGCCCCACGGACACCAUCCGUUCGGGAGUGCGCCGCAUCUG .....------...((.....(((((.((....(((((....)))))....)).)))))..((((((((((((.......)))).(((((.....)))))..)))).))))..))..... ( -44.00) >DroVir_CAF1 7367 114 - 1 CCACCGCC---GCCGCAUUUGCCGCCUGGUCA---UCCGCUGGGCAUCUUUCCAGCUGGCCCACAUCCGGGUCAUCCGCCGCCCCAUGGCCAUCAUCCAUUCGUGACUGCACCGCAUCUG .....((.---((.((....)).))..(((((---(..((((((......))))))((((((......))))))...((((.....))))............)))))).....))..... ( -32.50) >DroPse_CAF1 19628 111 - 1 CCGCC------CCCGCAUCUACCGCCCGGCCA---CCCAUUGGGCAUCUUUCCGGGCGGGCCACAUCCAGGACAUCCCCCACCCCAUGGCCAUCAUCCCUUCGGGGCGGCUCCUCAUCUC (((((------((.........(((((((...---(((...))).......)))))))(((((......((.......))......)))))...........)))))))........... ( -41.30) >DroMoj_CAF1 8709 117 - 1 CCGCCGCCACCACCGCAUCUGAUGCCCGGCCA---UCCGCUAGGCAUAUUUCCAGGCGGUCCCCAUCCAGGACAUCCACCGCCUCAUGGUCAUCAUCCAUUUGUGGCCGCUCCUCAUCUA ..((.(((((.(((((.....(((((..((..---...))..)))))........))))).........(((.....((((.....)))).....)))....))))).)).......... ( -35.32) >DroAna_CAF1 21307 114 - 1 CCACA------UCCGCAUCUACCGCCCGGCCAAAUACCGCUCGGCCUAUUCCCGGGCGGUCCGCAUCCCGUACAUCCAUCGCCCCAUGGACACCAUCCCUUCGGAGGACCUCCGCACCUG .....------...((.......((((((.......((....)).......))))))(((((............(((((......))))).....(((....))))))))...))..... ( -32.84) >DroPer_CAF1 19669 108 - 1 CCGCC------CCCGCAUCUGCCGCCCGGCCA---CCCAUUGGGCAUCUUUCCG-GCGGGCCACAUCCAGGGCAUCCCCCACCCCAUGGCCAUCAUCCCUUCGG-GCGGCU-CUCAUCUC ..((.------...))....(((((((((((.---.......)))........(-(..(((((......(((.....)))......))))).....))...)))-))))).-........ ( -36.60) >consensus CCACC______CCCGCAUCUGCCGCCCGGCCA___UCCACUGGGCAUCUUUCCGGGCGGGCCACAUCCAGGACAUCCACCGCCCCAUGGCCAUCAUCCCUUCGGGGCGGCUCCGCAUCUG ..............((.....((((((((.......((....)).......))))))))..........(((.....((((.....)))).....)))..........)).......... (-16.92 = -17.47 + 0.56)


| Location | 9,699,134 – 9,699,241 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.90 |
| Mean single sequence MFE | -34.73 |
| Consensus MFE | -20.50 |
| Energy contribution | -21.67 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9699134 107 - 27905053 -------CUUCCGCCCGUGCCAACUGGACCGCCACAUGGUCCACA---UCCGCAUCUGCCGCCCGGCCAAAUUCCACUUGGAAUCUUUCCCGGCGGACCGCAUCCCGGGCAUCCUCC -------.....((((((((....(((((((.....)))))))..---...)))....(((((.((....(((((....)))))....)).))))).........)))))....... ( -39.60) >DroVir_CAF1 7407 114 - 1 UCCGCAGCAUCCGCCCGUGCCAACGGGGCCACAUCAUGGUCCACCGCCGCCGCAUUUGCCGCCUGGUCA---UCCGCUGGGCAUCUUUCCAGCUGGCCCACAUCCGGGUCAUCCGCC ......((....(((((.(......((((((......((...((((..((.((....)).)).))))..---.))((((((......))))))))))))....)))))).....)). ( -38.20) >DroPse_CAF1 19668 108 - 1 UCCU---CAUCCGCCCGUGCCAACAGGACCGCCGCAUGGUCCGCC---CCCGCAUCUACCGCCCGGCCA---CCCAUUGGGCAUCUUUCCGGGCGGGCCACAUCCAGGACAUCCCCC ((((---.....((((((((.....((((((.....))))))...---...)))).....((((((...---(((...))).......)))))))))).......))))........ ( -36.80) >DroWil_CAF1 7556 99 - 1 UUCU---CAUCCACCCGUGCCAACUGGACCGCCACUUGGUCCUCC---UCCGCAUUUGCCACCUGGCCA---UCCACUU---AUGUUUCCCGGC------CAUCCAGCGCAUCCACC ....---.........((((.....((((((.....))))))...---...((....))....(((((.---...((..---..)).....)))------))....))))....... ( -21.70) >DroAna_CAF1 21347 107 - 1 -------CAUCCUCCCGUGCCAACUGGACCGCCCCACGGCCCACA---UCCGCAUCUACCGCCCGGCCAAAUACCGCUCGGCCUAUUCCCGGGCGGUCCGCAUCCCGUACAUCCAUC -------.........((((....((((((((((..(((......---.)))........(((.(((........))).)))........))))))))))......))))....... ( -33.70) >DroPer_CAF1 19707 107 - 1 UCCU---CAUCCGCCCGUGCCAACAGGACCGCCGCAUGGUCCGCC---CCCGCAUCUGCCGCCCGGCCA---CCCAUUGGGCAUCUUUCCG-GCGGGCCACAUCCAGGGCAUCCCCC ....---.....((((((((.....((....))))))((((((((---...((....)).((((((...---....))))))........)-))))))).......))))....... ( -38.40) >consensus UCCU___CAUCCGCCCGUGCCAACUGGACCGCCACAUGGUCCACC___UCCGCAUCUGCCGCCCGGCCA___UCCACUGGGCAUCUUUCCGGGCGGGCCACAUCCAGGGCAUCCACC ................(((......((((((.....))))))................((((((((.......((....)).......))))))))..)))................ (-20.50 = -21.67 + 1.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:19 2006