| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,394,175 – 1,394,276 |
| Length | 101 |
| Max. P | 0.580634 |

| Location | 1,394,175 – 1,394,276 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.84 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -18.83 |
| Energy contribution | -18.80 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
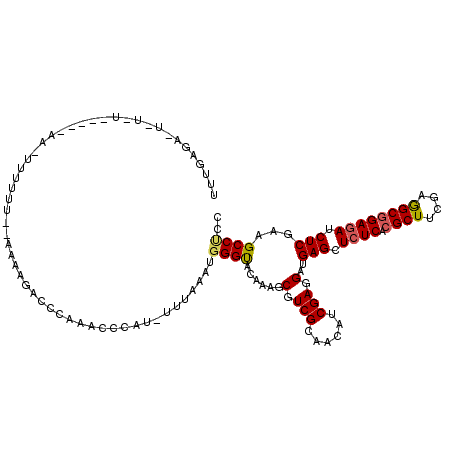
>3R_DroMel_CAF1 1394175 101 - 27905053 UUUGA----------------UUUUUU--AAAAGAGAAAACCCCAC-UUCAAAUGGGUACAAAGCGUCGCAACAUCGAGGAUGAGCUCUCACGCUUCGAGGCGGAGAUCUCGAAGCCCCC .....----------------......--....((((....((((.-......))))..((...(.(((......))).).))...))))..(((((((((......))))))))).... ( -23.70) >DroVir_CAF1 10411 116 - 1 UUUGAAUUUUUAUUGUUGACAUC---UCAACAAUAGCGUACGCCUU-UCGAAAUGGGCACAAAACGUCGCAAUAUCGAAGAUGAGCUCUCGCGCUUCGAAGCGGAGAUCUCUAAGCCGCC ..........(((((((((....---)))))))))(((...((.((-((((.((..((((.....)).)).)).))))))..(((.((((.((((....)))))))).)))...))))). ( -32.20) >DroSim_CAF1 10001 101 - 1 UUUGA----------------UUUUUU--AAAAGAGCCAAACCCAU-UUAAAAUGGGUACAAAGCGUCGCAACAUCGAGGAUGAGCUCUCACGCUUCGAGGCGGAGAUCUCGAAGCCUCC .....----------------......--...(((((((.((((((-.....))))))......(.(((......))).).)).)))))...(((((((((......))))))))).... ( -29.90) >DroEre_CAF1 10868 112 - 1 UUUGAGAAUCU-UC----CAGUUUUCU--CAUAGCCCCAAACCUAU-GUUAAAUGGGUACAAAGCGUCGCAACAUCGAGGAUGAGCUCUCUCGCUUCGAGGCGGAGAUCUCGAAGCCUCC ...(((...((-((----.((...(((--(...((.....((((((-.....)))))).....))..(((....((((((.((((....)))))))))).))))))).)).)))).))). ( -33.40) >DroYak_CAF1 9970 112 - 1 UCUGAGAAUCU-UC----AAGUUUUCU--AGUAGCCCCAAACCCAC-UUUAAAUGGGUACAAAGCGUCGGAACAUCGAGGAUGAGCUCUCACGCUUCGAGGCGGAGAUCUCGAAGCCUCC ((((((((...-..----.....))).--....((.....(((((.-......))))).....)).))))).....((((.((((.((((.((((....)))))))).))))...)))). ( -31.70) >DroPer_CAF1 12620 110 - 1 UUUGAGAGUUA-AA----AAAUUAUUU-----AUCCAUUAAGAUAUCAAUAAAUGGGUACGAAACGUCGCAAUAUCGAGGACGAGCUGUCUCGCUUCGAAGCUGAGAUCUCCAAGCCCCC ((((.(((...-..----........(-----(((((((............))))))))......(((.((.(.((((((.((((....)))))))))).).)).))))))))))..... ( -23.10) >consensus UUUGAGA_U_U_U_____AA_UUUUUU__AAAAGACCCAAACCCAU_UUUAAAUGGGUACAAAGCGUCGCAACAUCGAGGAUGAGCUCUCACGCUUCGAGGCGGAGAUCUCGAAGCCUCC ......................................................((((......(.(((......))).)..(((.((((.((((....)))))))).)))...)))).. (-18.83 = -18.80 + -0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:13 2006