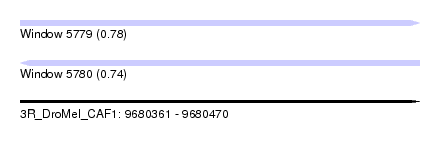
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,680,361 – 9,680,470 |
| Length | 109 |
| Max. P | 0.783636 |

| Location | 9,680,361 – 9,680,470 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.79 |
| Mean single sequence MFE | -29.81 |
| Consensus MFE | -19.78 |
| Energy contribution | -20.87 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
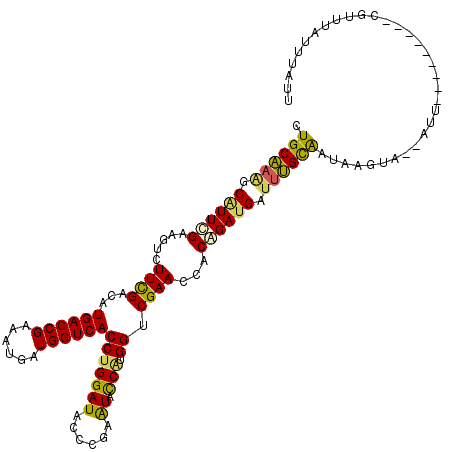
>3R_DroMel_CAF1 9680361 109 + 27905053 AAUAAAUAAACG---------AAU--UACUUAUUACAAAUCAUCUGUGGUUCGACCAUGGUAUUCGGGUAUCCAGGUGACCAUCAUUUCGGUCAUGUCGAAGACUGCGAAUGCUUUGCAG ..........((---------(((--(((................))))))))..((.(((((((((((.((.(.((((((........)))))).).))..))).)))))))).))... ( -28.79) >DroSec_CAF1 4593 109 + 1 AAUAAAUAAACG---------AAU--UACUUAUUGCAAAUCAUCUGUGGUUCGACCAUGGUAUUCGGGUAUCCAGGUGACCACCAUUUCGGUCAUAUCGAAAACUGCAAAUGCUUUGCAG ............---------...--.......((((((.(((.((..((((((((.((((((....))).)))))(((((........)))))..))))..))..)).))).)))))). ( -28.50) >DroSim_CAF1 3670 109 + 1 AAUAAAUAAACG---------AAU--UACUUAUUGCAAAUCAUCUGUGGUUCGACCAUGGUAUUCGGGUAUCCAGGUGACCAUCAUUUCGGUCAUGUCGAAGACUGCAAAUGCUUUGCAG ............---------...--.......((((((.(((.((..(((((((..((((((....))).))).((((((........)))))))))))..))..)).))).)))))). ( -30.80) >DroEre_CAF1 4382 118 + 1 AAAAAAUACAUCAAAUUGGGCAAU--UACUUAUUGCAAAUCAUCGGUAGUUCGGCCAUGGAACUCGGGUAUCCAGGUGACCAUCAUUUCGGUCAUGUCGAACACCUCCAACGCUUUGCAA ...............(((((((((--.....)))))........(((.(((((((..((((((....)).)))).((((((........)))))))))))))))).)))).......... ( -33.00) >DroYak_CAF1 3875 118 + 1 GAUAAAUAAAUAAAAUUGGGCAAU--UACUUAUUGCAACUCAUCGGUAGUUCGGCCGUGGAACUCGGGUAUCCUGGUGACCAUCAUUUCGGUCAUGUCAAAGCACUCCAACGCUUUGCAG ...................(((((--.....)))))...(((.((((......)))))))....((((...))))((((((........))))))(.((((((........))))))).. ( -27.20) >DroAna_CAF1 3637 103 + 1 -----------------GGGAAAUCUAAUGUGCCGCGUAUCUUCGGAGGUUCGUCCACGAUACUCGGGUAUCCAGGUGACCAUCAUUUCGGUCAUGUCGAAGACCUCGAACGCCUCGCAG -----------------...........((((..((((....(((.(((((..((.(((((((....)))))...((((((........)))))))).)).))))))))))))..)))). ( -30.60) >consensus AAUAAAUAAACG_________AAU__UACUUAUUGCAAAUCAUCGGUGGUUCGACCAUGGUACUCGGGUAUCCAGGUGACCAUCAUUUCGGUCAUGUCGAAGACCGCAAACGCUUUGCAG ................................(((((((.(...(((((((((((...(((((....)))))...((((((........)))))))))))..))))))...).))))))) (-19.78 = -20.87 + 1.09)



| Location | 9,680,361 – 9,680,470 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.79 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -20.89 |
| Energy contribution | -20.20 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.735027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9680361 109 - 27905053 CUGCAAAGCAUUCGCAGUCUUCGACAUGACCGAAAUGAUGGUCACCUGGAUACCCGAAUACCAUGGUCGAACCACAGAUGAUUUGUAAUAAGUA--AUU---------CGUUUAUUUAUU .((((((.(((..(..((..(((((.((((((......))))))..(((....))).........)))))))..)..))).))))))(((((((--(..---------...)))))))). ( -24.20) >DroSec_CAF1 4593 109 - 1 CUGCAAAGCAUUUGCAGUUUUCGAUAUGACCGAAAUGGUGGUCACCUGGAUACCCGAAUACCAUGGUCGAACCACAGAUGAUUUGCAAUAAGUA--AUU---------CGUUUAUUUAUU .((((((.((((((..((..((((..((((((......))))))(((((((......)).))).))))))))..)))))).))))))(((((((--(..---------...)))))))). ( -28.60) >DroSim_CAF1 3670 109 - 1 CUGCAAAGCAUUUGCAGUCUUCGACAUGACCGAAAUGAUGGUCACCUGGAUACCCGAAUACCAUGGUCGAACCACAGAUGAUUUGCAAUAAGUA--AUU---------CGUUUAUUUAUU .((((((.((((((..((..(((((.((((((......))))))..(((....))).........)))))))..)))))).))))))(((((((--(..---------...)))))))). ( -29.80) >DroEre_CAF1 4382 118 - 1 UUGCAAAGCGUUGGAGGUGUUCGACAUGACCGAAAUGAUGGUCACCUGGAUACCCGAGUUCCAUGGCCGAACUACCGAUGAUUUGCAAUAAGUA--AUUGCCCAAUUUGAUGUAUUUUUU (((((((.((((((.((((((((...((((((......))))))..))))))))..(((((.......))))).)))))).)))))))......--........................ ( -33.50) >DroYak_CAF1 3875 118 - 1 CUGCAAAGCGUUGGAGUGCUUUGACAUGACCGAAAUGAUGGUCACCAGGAUACCCGAGUUCCACGGCCGAACUACCGAUGAGUUGCAAUAAGUA--AUUGCCCAAUUUUAUUUAUUUAUC ..((((..((((((((((((.((...((((((......))))))...((....))......)).)))...))).))))))..))))((((((((--(..........))))))))).... ( -29.40) >DroAna_CAF1 3637 103 - 1 CUGCGAGGCGUUCGAGGUCUUCGACAUGACCGAAAUGAUGGUCACCUGGAUACCCGAGUAUCGUGGACGAACCUCCGAAGAUACGCGGCACAUUAGAUUUCCC----------------- (((((...(.(((((((((.((.((.((((((......))))))....(((((....))))))).)).).)))).)))))...)))))...............----------------- ( -29.70) >consensus CUGCAAAGCAUUCGAAGUCUUCGACAUGACCGAAAUGAUGGUCACCUGGAUACCCGAAUACCAUGGUCGAACCACAGAUGAUUUGCAAUAAGUA__AUU_________CGUUUAUUUAUU .((((((.((((((.....((((...((((((......))))))(((((((......)).))).)).))))...)))))).))))))................................. (-20.89 = -20.20 + -0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:09 2006