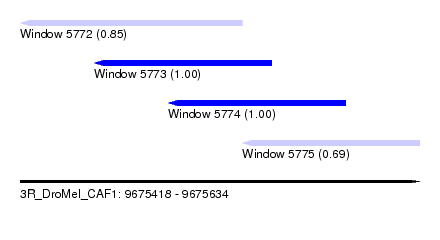
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,675,418 – 9,675,634 |
| Length | 216 |
| Max. P | 0.999865 |

| Location | 9,675,418 – 9,675,538 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -27.64 |
| Consensus MFE | -25.94 |
| Energy contribution | -26.22 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9675418 120 - 27905053 CCACAUUCACGUCCACAUCCAUGGGCGAUGGGCAUGAAAAUCGUUUACAAAUAAAUCAGAAAGCGAUAAAUAAGGAUUUUGUCUCGUAAUAACUCAGCGGGAGCCAUUUAACAUGCAAUC ......(((((((((......)))))).)))(((((...(((((((.(..........).)))))))......((......((((((.........)))))).))......))))).... ( -26.60) >DroSec_CAF1 67151 120 - 1 CCACAUUCACGUCCACGUCCAUGGGCGAUGGGCAUGAAAAUCGUUUACAAAUAAAUCAGAAAGCGAUAAAUAAGGAUUUUGUCUCGUAAUAACUCAGCGGGAGCCAUUUAACAUGCAAUC ((...((((.(((((((((....)))).))))).)))).(((((((.(..........).)))))))......))......((((((.........)))))).................. ( -27.90) >DroSim_CAF1 70078 120 - 1 CCACAUUCACGUCCACGUCCAUGGGCGAUGGGCAUGAAAAUCGUUUACAAAUAAAUCAGAAAGCGAUAAAUAAGGAUUUUGUCUCGUAAUAACUCAGCGGGAGCCAUUUAACAUGCAAUC ((...((((.(((((((((....)))).))))).)))).(((((((.(..........).)))))))......))......((((((.........)))))).................. ( -27.90) >DroEre_CAF1 68374 112 - 1 --------ACGUCCACGUCCAUGGGCGAUGGGCAUGAAAAUCGUUUACAAAUAAAUCAGAAAGCGAUAAAUAAGGAUUUUGUCUCGUAAUAACUCAGCGGGAGCCAUUUAACAUGCAAUC --------....(((((((....)))).)))(((((...(((((((.(..........).)))))))......((......((((((.........)))))).))......))))).... ( -26.50) >DroYak_CAF1 72209 120 - 1 UCACAUCCACGUCCACGUCCAUGGGCGAUGGGCAUGAAAAUCGUUUACAAAUAAAUCAGAAAGCGAUAAAUAAGGAUUUUGUCUCGUAAUAACUCAGCGGGAGCCAUUUAACAUGCAAUC ......(((((((((......)))))).)))(((((...(((((((.(..........).)))))))......((......((((((.........)))))).))......))))).... ( -29.30) >consensus CCACAUUCACGUCCACGUCCAUGGGCGAUGGGCAUGAAAAUCGUUUACAAAUAAAUCAGAAAGCGAUAAAUAAGGAUUUUGUCUCGUAAUAACUCAGCGGGAGCCAUUUAACAUGCAAUC ......(((((((((......)))))).)))(((((...(((((((.(..........).)))))))......((......((((((.........)))))).))......))))).... (-25.94 = -26.22 + 0.28)



| Location | 9,675,458 – 9,675,554 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.33 |
| Mean single sequence MFE | -23.43 |
| Consensus MFE | -21.54 |
| Energy contribution | -21.85 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.999252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9675458 96 - 27905053 ACAUC------------CACAUC------------CACAUCCACAUUCACGUCCACAUCCAUGGGCGAUGGGCAUGAAAAUCGUUUACAAAUAAAUCAGAAAGCGAUAAAUAAGGAUUUU .....------------...(((------------(.(((((.(((...((((((......))))))))))).)))...(((((((.(..........).)))))))......))))... ( -20.50) >DroSec_CAF1 67191 84 - 1 AC------------------------------------AUCCACAUUCACGUCCACGUCCAUGGGCGAUGGGCAUGAAAAUCGUUUACAAAUAAAUCAGAAAGCGAUAAAUAAGGAUUUU ..------------------------------------((((...((((.(((((((((....)))).))))).)))).(((((((.(..........).)))))))......))))... ( -24.70) >DroSim_CAF1 70118 96 - 1 ACAUC------------CACAUC------------CACAUCCACAUUCACGUCCACGUCCAUGGGCGAUGGGCAUGAAAAUCGUUUACAAAUAAAUCAGAAAGCGAUAAAUAAGGAUUUU .....------------......------------...((((...((((.(((((((((....)))).))))).)))).(((((((.(..........).)))))))......))))... ( -24.70) >DroYak_CAF1 72249 120 - 1 ACAUCCCCAUCCCCAUCUCCAUCCACAACUACAUCCACAUUCACAUCCACGUCCACGUCCAUGGGCGAUGGGCAUGAAAAUCGUUUACAAAUAAAUCAGAAAGCGAUAAAUAAGGAUUUU ................................((((...((((..((((((((((......)))))).))))..)))).(((((((.(..........).)))))))......))))... ( -23.80) >consensus ACAUC____________CACAUC____________CACAUCCACAUUCACGUCCACGUCCAUGGGCGAUGGGCAUGAAAAUCGUUUACAAAUAAAUCAGAAAGCGAUAAAUAAGGAUUUU ......................................((((...((((.(((((((((....)))).))))).)))).(((((((.(..........).)))))))......))))... (-21.54 = -21.85 + 0.31)

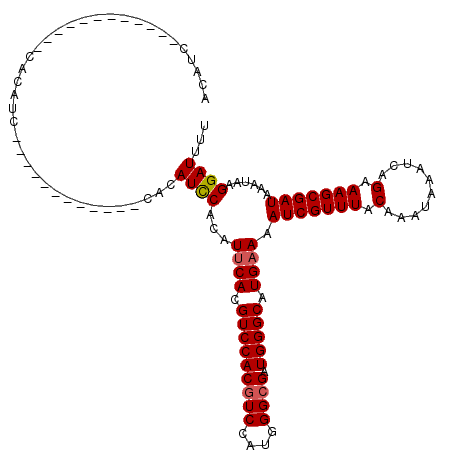
| Location | 9,675,498 – 9,675,594 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.62 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -23.41 |
| Energy contribution | -24.66 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.43 |
| Structure conservation index | 0.85 |
| SVM decision value | 4.30 |
| SVM RNA-class probability | 0.999865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9675498 96 - 27905053 GUCCUGUUGGCCAUACUCAGCCACAUCUUGGCCAAGAACCACAUC------------CACAUC------------CACAUCCACAUUCACGUCCACAUCCAUGGGCGAUGGGCAUGAAAA (((((.(((((((...............))))))).)........------------......------------..............((((((......))))))..))))....... ( -23.76) >DroSec_CAF1 67231 84 - 1 GUCCUGUUGGCCAUCCUCAGCCACAGCUUGGCCAACAUCCAC------------------------------------AUCCACAUUCACGUCCACGUCCAUGGGCGAUGGGCAUGAAAA ....(((((((((.....(((....)))))))))))).....------------------------------------.......((((.(((((((((....)))).))))).)))).. ( -31.90) >DroSim_CAF1 70158 96 - 1 GUCCUGUUGGCCAUCCUCAGCCACAUCUUGGCCAACAUCCACAUC------------CACAUC------------CACAUCCACAUUCACGUCCACGUCCAUGGGCGAUGGGCAUGAAAA ....(((((((((...............)))))))))........------------......------------..........((((.(((((((((....)))).))))).)))).. ( -30.16) >DroYak_CAF1 72289 120 - 1 GUCCUGUUGGCCAUCCUCAUCCUCAUCUUGGGCCACAUCCACAUCCCCAUCCCCAUCUCCAUCCACAACUACAUCCACAUUCACAUCCACGUCCACGUCCAUGGGCGAUGGGCAUGAAAA ....(((.((((..................)))))))....(((.(((((((((((............................................))))).)))))).))).... ( -24.27) >consensus GUCCUGUUGGCCAUCCUCAGCCACAUCUUGGCCAACAUCCACAUC____________CACAUC____________CACAUCCACAUUCACGUCCACGUCCAUGGGCGAUGGGCAUGAAAA ....(((((((((...............)))))))))................................................((((.(((((((((....)))).))))).)))).. (-23.41 = -24.66 + 1.25)


| Location | 9,675,538 – 9,675,634 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.09 |
| Mean single sequence MFE | -22.03 |
| Consensus MFE | -19.75 |
| Energy contribution | -19.50 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9675538 96 - 27905053 UGGCCAGCCCAUCAAGAUGCCAGUCGAAGUCCAGCUGCAGGUCCUGUUGGCCAUACUCAGCCACAUCUUGGCCAAGAACCACAUC------------CACAUC------------CACAU ((((((((....(....(((.(((.(.....).)))))).)....))))))))..((..((((.....))))..)).........------------......------------..... ( -22.80) >DroSec_CAF1 67271 84 - 1 UGGCCAGCCCAUCAAGAUGCCAGACGAAGUCCAGCUGCAGGUCCUGUUGGCCAUCCUCAGCCACAGCUUGGCCAACAUCCAC------------------------------------AU ..((.(((.(((....)))...(((...)))..))))).((...(((((((((.....(((....)))))))))))).))..------------------------------------.. ( -22.50) >DroSim_CAF1 70198 96 - 1 UGGCCAGCCCAUCAAGAUGCCAGUCGAAGUCCAGCUGCAGGUCCUGUUGGCCAUCCUCAGCCACAUCUUGGCCAACAUCCACAUC------------CACAUC------------CACAU (((.((((.......(((....)))........))))..((...(((((((((...............))))))))).))....)------------))....------------..... ( -22.32) >DroYak_CAF1 72329 120 - 1 UGGCCAGCCCAUCAAGAUGCCAGUCGAAGUCCAACUGCAGGUCCUGUUGGCCAUCCUCAUCCUCAUCUUGGGCCACAUCCACAUCCCCAUCCCCAUCUCCAUCCACAACUACAUCCACAU (((((.......(((((((.((((.(.....).))))..((.((....))))...........))))))))))))............................................. ( -20.51) >consensus UGGCCAGCCCAUCAAGAUGCCAGUCGAAGUCCAGCUGCAGGUCCUGUUGGCCAUCCUCAGCCACAUCUUGGCCAACAUCCACAUC____________CACAUC____________CACAU ((((((((....(....(((.(((.(.....).)))))).)....))))))))......(((.......)))................................................ (-19.75 = -19.50 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:04 2006