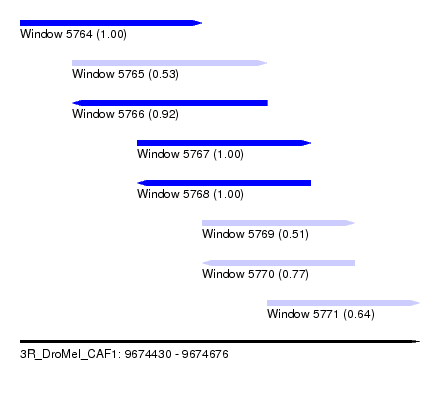
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,674,430 – 9,674,676 |
| Length | 246 |
| Max. P | 0.999324 |

| Location | 9,674,430 – 9,674,542 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.20 |
| Mean single sequence MFE | -37.57 |
| Consensus MFE | -36.09 |
| Energy contribution | -36.09 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.51 |
| SVM RNA-class probability | 0.999324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9674430 112 + 27905053 GCCAUCGGGCCAUC--------GAGCCAGCUCGUUUCCUGUGACACAGUUCUGGCAAUUAGUCAUGGGAAUCACGUAUACGCCCUGCGUGUCUGGCAGCCAUUUUAUGGCAAAAGGCCUC ......(((((...--------..(((((..(((((((((((((................))))))))))..))).((((((...))))))))))).(((((...)))))....))))). ( -40.59) >DroSec_CAF1 66175 104 + 1 --------GCCAUC--------GAGCCAGCUCGUUUCCUGUGACACAGUUCUGGCAAUUAGUCAUGGGAAUCACGUAUACGCCCUGCGUGUCUGGCAGCCAUUUUAUGGCAAAAGGCCUC --------(((...--------..(((((..(((((((((((((................))))))))))..))).((((((...))))))))))).(((((...)))))....)))... ( -36.09) >DroSim_CAF1 69113 104 + 1 --------GCCAUC--------GAGCCAGCUCGUUUCCUGUGACACAGUUCUGGCAAUUAGUCAUGGGAAUCACGUAUACGCCCUGCGUGUCUGGCAGCCAUUUUAUGGCAAAAGGCCUC --------(((...--------..(((((..(((((((((((((................))))))))))..))).((((((...))))))))))).(((((...)))))....)))... ( -36.09) >DroEre_CAF1 67327 112 + 1 --------GCCAUCGAGCCAUCGAGCCAGCUCGUUUCCUGUGACACAGUUCUGGCAAUUAGUCAUGGGAAUCACGUAUACGCCCUGCGUGUCUGGCAGCCAUUUUAUGGCAAAAGGCCUC --------(((.((((....))))(((((..(((((((((((((................))))))))))..))).((((((...))))))))))).(((((...)))))....)))... ( -38.99) >DroYak_CAF1 71163 104 + 1 --------GCCAUC--------GAGCCAGCUCGUUUCCUGUGACACAGUUCUGGCAAUUAGUCAUGGGAAUCACGUAUACGCCCUGCGUGUCUGGCAGCCAUUUUAUGGCAAAAGGCCUC --------(((...--------..(((((..(((((((((((((................))))))))))..))).((((((...))))))))))).(((((...)))))....)))... ( -36.09) >consensus ________GCCAUC________GAGCCAGCUCGUUUCCUGUGACACAGUUCUGGCAAUUAGUCAUGGGAAUCACGUAUACGCCCUGCGUGUCUGGCAGCCAUUUUAUGGCAAAAGGCCUC ........(((.............(((((..(((((((((((((................))))))))))..))).((((((...))))))))))).(((((...)))))....)))... (-36.09 = -36.09 + -0.00)

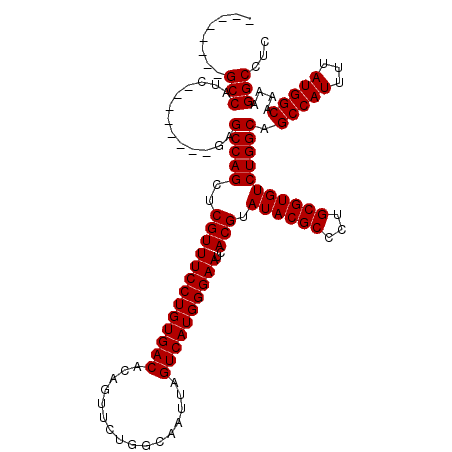
| Location | 9,674,462 – 9,674,582 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.67 |
| Mean single sequence MFE | -33.24 |
| Consensus MFE | -32.32 |
| Energy contribution | -32.32 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.97 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9674462 120 + 27905053 UGACACAGUUCUGGCAAUUAGUCAUGGGAAUCACGUAUACGCCCUGCGUGUCUGGCAGCCAUUUUAUGGCAAAAGGCCUCAACGUCAACGCUGGGCUUUUGUAAUUUUAAUUACUUCCCC ...........((((.....)))).(((((..........((((.((((....(((.(((((...))))).....))).........)))).))))....(((((....)))))))))). ( -32.32) >DroSec_CAF1 66199 120 + 1 UGACACAGUUCUGGCAAUUAGUCAUGGGAAUCACGUAUACGCCCUGCGUGUCUGGCAGCCAUUUUAUGGCAAAAGGCCUCAACGUCAACGCAGGGCUUUUGUAAUUUUAAUUACUUCCCC ...........((((.....)))).(((((..........(((((((((....(((.(((((...))))).....))).........)))))))))....(((((....)))))))))). ( -36.92) >DroSim_CAF1 69137 120 + 1 UGACACAGUUCUGGCAAUUAGUCAUGGGAAUCACGUAUACGCCCUGCGUGUCUGGCAGCCAUUUUAUGGCAAAAGGCCUCAACGUCAACGCUGGGCUUUUGUAAUUUUAAUUACUUCCCC ...........((((.....)))).(((((..........((((.((((....(((.(((((...))))).....))).........)))).))))....(((((....)))))))))). ( -32.32) >DroEre_CAF1 67359 120 + 1 UGACACAGUUCUGGCAAUUAGUCAUGGGAAUCACGUAUACGCCCUGCGUGUCUGGCAGCCAUUUUAUGGCAAAAGGCCUCAACGUCAACGCUGGGCUUUUGUAAUUUUAAUUACUUCCCC ...........((((.....)))).(((((..........((((.((((....(((.(((((...))))).....))).........)))).))))....(((((....)))))))))). ( -32.32) >DroYak_CAF1 71187 120 + 1 UGACACAGUUCUGGCAAUUAGUCAUGGGAAUCACGUAUACGCCCUGCGUGUCUGGCAGCCAUUUUAUGGCAAAAGGCCUCAACGUCAACGCUGGGCUUUUGUAAUUUUAAUUACUUCCCC ...........((((.....)))).(((((..........((((.((((....(((.(((((...))))).....))).........)))).))))....(((((....)))))))))). ( -32.32) >consensus UGACACAGUUCUGGCAAUUAGUCAUGGGAAUCACGUAUACGCCCUGCGUGUCUGGCAGCCAUUUUAUGGCAAAAGGCCUCAACGUCAACGCUGGGCUUUUGUAAUUUUAAUUACUUCCCC ...........((((.....)))).(((((..........((((.((((....(((.(((((...))))).....))).........)))).))))....(((((....)))))))))). (-32.32 = -32.32 + 0.00)

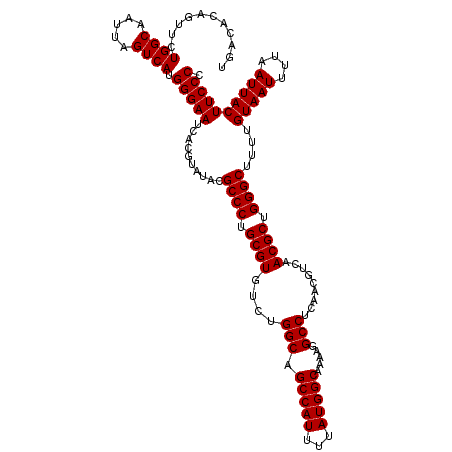
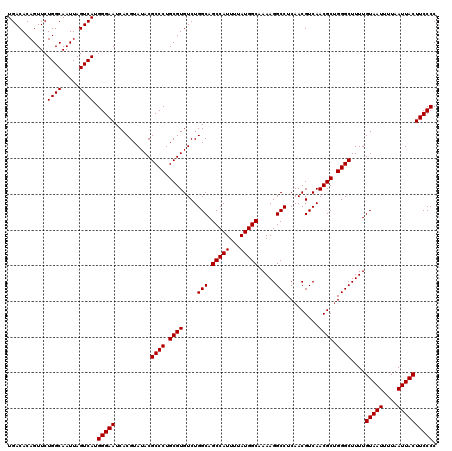
| Location | 9,674,462 – 9,674,582 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.67 |
| Mean single sequence MFE | -35.84 |
| Consensus MFE | -34.83 |
| Energy contribution | -34.83 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9674462 120 - 27905053 GGGGAAGUAAUUAAAAUUACAAAAGCCCAGCGUUGACGUUGAGGCCUUUUGCCAUAAAAUGGCUGCCAGACACGCAGGGCGUAUACGUGAUUCCCAUGACUAAUUGCCAGAACUGUGUCA .((...(((((....)))))((((((((((((....))))).)).)))))(((((...)))))..)).((((((...((((....((((.....))))......)))).....)))))). ( -34.80) >DroSec_CAF1 66199 120 - 1 GGGGAAGUAAUUAAAAUUACAAAAGCCCUGCGUUGACGUUGAGGCCUUUUGCCAUAAAAUGGCUGCCAGACACGCAGGGCGUAUACGUGAUUCCCAUGACUAAUUGCCAGAACUGUGUCA .((((((((((....)))))....(((((((((....(((..(((.....(((((...))))).))).))))))))))))..........))))).((((................)))) ( -39.99) >DroSim_CAF1 69137 120 - 1 GGGGAAGUAAUUAAAAUUACAAAAGCCCAGCGUUGACGUUGAGGCCUUUUGCCAUAAAAUGGCUGCCAGACACGCAGGGCGUAUACGUGAUUCCCAUGACUAAUUGCCAGAACUGUGUCA .((...(((((....)))))((((((((((((....))))).)).)))))(((((...)))))..)).((((((...((((....((((.....))))......)))).....)))))). ( -34.80) >DroEre_CAF1 67359 120 - 1 GGGGAAGUAAUUAAAAUUACAAAAGCCCAGCGUUGACGUUGAGGCCUUUUGCCAUAAAAUGGCUGCCAGACACGCAGGGCGUAUACGUGAUUCCCAUGACUAAUUGCCAGAACUGUGUCA .((...(((((....)))))((((((((((((....))))).)).)))))(((((...)))))..)).((((((...((((....((((.....))))......)))).....)))))). ( -34.80) >DroYak_CAF1 71187 120 - 1 GGGGAAGUAAUUAAAAUUACAAAAGCCCAGCGUUGACGUUGAGGCCUUUUGCCAUAAAAUGGCUGCCAGACACGCAGGGCGUAUACGUGAUUCCCAUGACUAAUUGCCAGAACUGUGUCA .((...(((((....)))))((((((((((((....))))).)).)))))(((((...)))))..)).((((((...((((....((((.....))))......)))).....)))))). ( -34.80) >consensus GGGGAAGUAAUUAAAAUUACAAAAGCCCAGCGUUGACGUUGAGGCCUUUUGCCAUAAAAUGGCUGCCAGACACGCAGGGCGUAUACGUGAUUCCCAUGACUAAUUGCCAGAACUGUGUCA .((((((((((....)))))....((((.((((....(((..(((.....(((((...))))).))).))))))).))))..........))))).((((................)))) (-34.83 = -34.83 + -0.00)

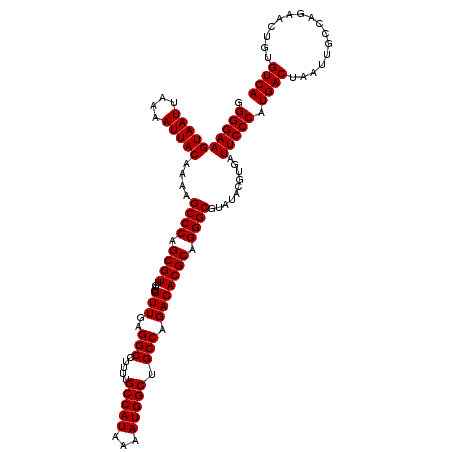
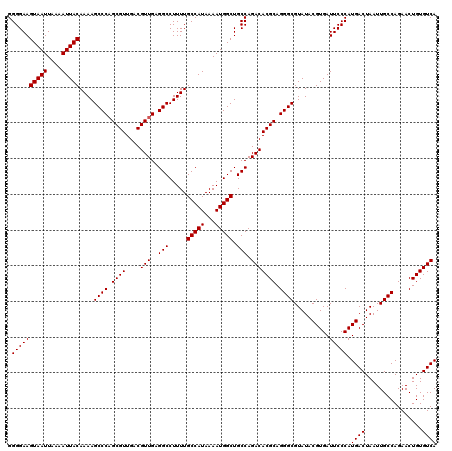
| Location | 9,674,502 – 9,674,609 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.94 |
| Mean single sequence MFE | -37.56 |
| Consensus MFE | -33.92 |
| Energy contribution | -33.76 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.998018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9674502 107 + 27905053 GCCCUGCGUGUCUGGCAGCCAUUUUAUGGCAAAAGGCCUCAACGUCAACGCUGGGCUUUUGUAAUUUUAAUUACUUCCCCUUGAUGGCUGGCUGAGUUAUUGGCCAG------------- ((((.((((....(((.(((((...))))).....))).........)))).))))....(((((....)))))....((.....))(((((..(....)..)))))------------- ( -34.02) >DroSec_CAF1 66239 107 + 1 GCCCUGCGUGUCUGGCAGCCAUUUUAUGGCAAAAGGCCUCAACGUCAACGCAGGGCUUUUGUAAUUUUAAUUACUUCCCCUUGUUGGCUGGCUGAGUUAUUAGCCAG------------- (((((((((....(((.(((((...))))).....))).........)))))))))....(((((....)))))....((.....))((((((((....))))))))------------- ( -39.52) >DroSim_CAF1 69177 107 + 1 GCCCUGCGUGUCUGGCAGCCAUUUUAUGGCAAAAGGCCUCAACGUCAACGCUGGGCUUUUGUAAUUUUAAUUACUUCCCCUUGAUGGCUGGCUGAGUUAUUGGCCAG------------- ((((.((((....(((.(((((...))))).....))).........)))).))))....(((((....)))))....((.....))(((((..(....)..)))))------------- ( -34.02) >DroEre_CAF1 67399 107 + 1 GCCCUGCGUGUCUGGCAGCCAUUUUAUGGCAAAAGGCCUCAACGUCAACGCUGGGCUUUUGUAAUUUUAAUUACUUCCCCCUGAUGGCUGGCUGAGUUAUUGGCCAG------------- ((((.((((....(((.(((((...))))).....))).........)))).))))....(((((....))))).....((....))(((((..(....)..)))))------------- ( -34.02) >DroYak_CAF1 71227 120 + 1 GCCCUGCGUGUCUGGCAGCCAUUUUAUGGCAAAAGGCCUCAACGUCAACGCUGGGCUUUUGUAAUUUUAAUUACUUCCCCUUGAUGGCUGGCUGAGUUAUUGGCCAGUGGGAGUGGGAGG ((((.((((....(((.(((((...))))).....))).........)))).)))).................(((((((((..(.((((((..(....)..)))))).)))).)))))) ( -46.22) >consensus GCCCUGCGUGUCUGGCAGCCAUUUUAUGGCAAAAGGCCUCAACGUCAACGCUGGGCUUUUGUAAUUUUAAUUACUUCCCCUUGAUGGCUGGCUGAGUUAUUGGCCAG_____________ ((((.((((....(((.(((((...))))).....))).........)))).))))....(((((....)))))....((.....))((((((((....))))))))............. (-33.92 = -33.76 + -0.16)

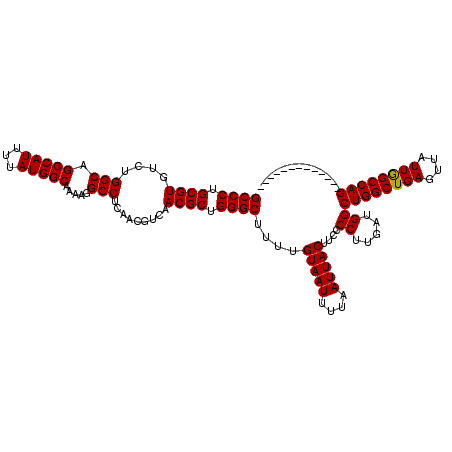

| Location | 9,674,502 – 9,674,609 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.94 |
| Mean single sequence MFE | -33.44 |
| Consensus MFE | -30.88 |
| Energy contribution | -30.88 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.30 |
| SVM RNA-class probability | 0.998962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9674502 107 - 27905053 -------------CUGGCCAAUAACUCAGCCAGCCAUCAAGGGGAAGUAAUUAAAAUUACAAAAGCCCAGCGUUGACGUUGAGGCCUUUUGCCAUAAAAUGGCUGCCAGACACGCAGGGC -------------...(((......((.(.(((((((....((.(((((((....)))))....((((((((....))))).)))...)).)).....))))))).).)).......))) ( -30.62) >DroSec_CAF1 66239 107 - 1 -------------CUGGCUAAUAACUCAGCCAGCCAACAAGGGGAAGUAAUUAAAAUUACAAAAGCCCUGCGUUGACGUUGAGGCCUUUUGCCAUAAAAUGGCUGCCAGACACGCAGGGC -------------((((((........))))))((......))...(((((....)))))....(((((((((....(((..(((.....(((((...))))).))).)))))))))))) ( -38.10) >DroSim_CAF1 69177 107 - 1 -------------CUGGCCAAUAACUCAGCCAGCCAUCAAGGGGAAGUAAUUAAAAUUACAAAAGCCCAGCGUUGACGUUGAGGCCUUUUGCCAUAAAAUGGCUGCCAGACACGCAGGGC -------------...(((......((.(.(((((((....((.(((((((....)))))....((((((((....))))).)))...)).)).....))))))).).)).......))) ( -30.62) >DroEre_CAF1 67399 107 - 1 -------------CUGGCCAAUAACUCAGCCAGCCAUCAGGGGGAAGUAAUUAAAAUUACAAAAGCCCAGCGUUGACGUUGAGGCCUUUUGCCAUAAAAUGGCUGCCAGACACGCAGGGC -------------...(((......((.(.(((((((...((.((((((((....)))))....((((((((....))))).)))..))).)).....))))))).).)).......))) ( -31.52) >DroYak_CAF1 71227 120 - 1 CCUCCCACUCCCACUGGCCAAUAACUCAGCCAGCCAUCAAGGGGAAGUAAUUAAAAUUACAAAAGCCCAGCGUUGACGUUGAGGCCUUUUGCCAUAAAAUGGCUGCCAGACACGCAGGGC ..((((.......(((((..........)))))........)))).(((((....)))))....((((.((((....(((..(((.....(((((...))))).))).))))))).)))) ( -36.36) >consensus _____________CUGGCCAAUAACUCAGCCAGCCAUCAAGGGGAAGUAAUUAAAAUUACAAAAGCCCAGCGUUGACGUUGAGGCCUUUUGCCAUAAAAUGGCUGCCAGACACGCAGGGC .............(((((..........)))))((......))...(((((....)))))....((((.((((....(((..(((.....(((((...))))).))).))))))).)))) (-30.88 = -30.88 + 0.00)

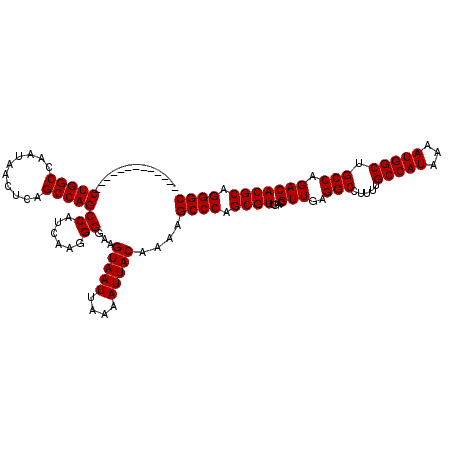
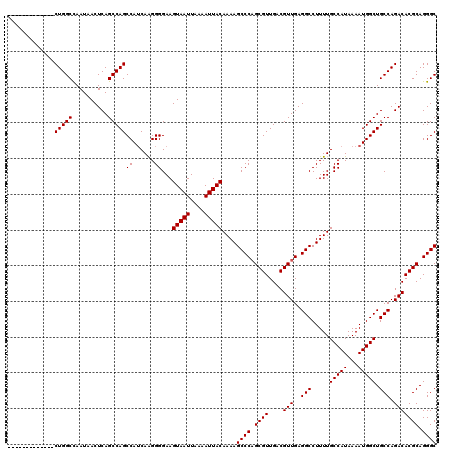
| Location | 9,674,542 – 9,674,636 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.08 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -21.59 |
| Energy contribution | -21.83 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9674542 94 + 27905053 AACGUCAACGCUGGGCUUUUGUAAUUUUAAUUACUUCCCCUUGAUGGCUGGCUGAGUUAUUGGCCAG--------------------AAAGUGGGA------GUGGGCGUAAGGAAGUCG .............((((((..((.......((((((((.(((.....(((((..(....)..)))))--------------------.))).))))------))))...))..)))))). ( -28.90) >DroSec_CAF1 66279 94 + 1 AACGUCAACGCAGGGCUUUUGUAAUUUUAAUUACUUCCCCUUGUUGGCUGGCUGAGUUAUUAGCCAG--------------------AAAGUGGGA------GUGGGCGUAAGGAAGUCG .(((((.((...(((.....(((((....)))))..)))((..((..((((((((....))))))))--------------------..))..)).------)).))))).......... ( -30.00) >DroSim_CAF1 69217 94 + 1 AACGUCAACGCUGGGCUUUUGUAAUUUUAAUUACUUCCCCUUGAUGGCUGGCUGAGUUAUUGGCCAG--------------------AAAGUGGGA------GUGGGCGUAAGGAAGUCA .............((((((..((.......((((((((.(((.....(((((..(....)..)))))--------------------.))).))))------))))...))..)))))). ( -28.90) >DroEre_CAF1 67439 98 + 1 AACGUCAACGCUGGGCUUUUGUAAUUUUAAUUACUUCCCCCUGAUGGCUGGCUGAGUUAUUGGCCAG--------------------AAAGUGGGAGUGG--GUGGGAGUAAUGAAGUCG .............((((((..........(((((((((((((.((..(((((..(....)..)))))--------------------...)).)....))--).))))))))))))))). ( -30.70) >DroYak_CAF1 71267 120 + 1 AACGUCAACGCUGGGCUUUUGUAAUUUUAAUUACUUCCCCUUGAUGGCUGGCUGAGUUAUUGGCCAGUGGGAGUGGGAGGUGCAGUGGAAGUGGGAGUGGGAGUGGGUGUAAGGAAGUCG .((.((.(((((.(.((((..(.(((((...(((((((((((..(.((((((..(....)..)))))).)))).)))))))).....)))))....)..))))).)))))...)).)).. ( -38.60) >consensus AACGUCAACGCUGGGCUUUUGUAAUUUUAAUUACUUCCCCUUGAUGGCUGGCUGAGUUAUUGGCCAG____________________AAAGUGGGA______GUGGGCGUAAGGAAGUCG .............((((((((((((....))))).(((((((.....((((((((....)))))))).....................))).))))................))))))). (-21.59 = -21.83 + 0.24)

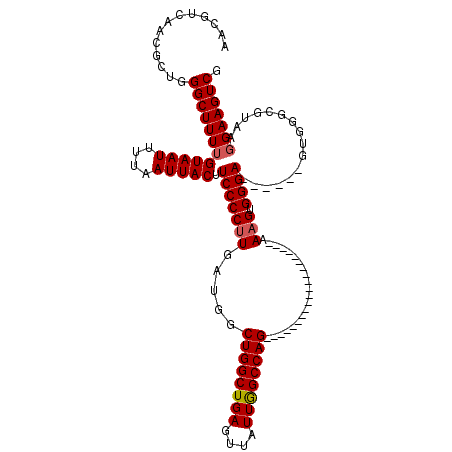
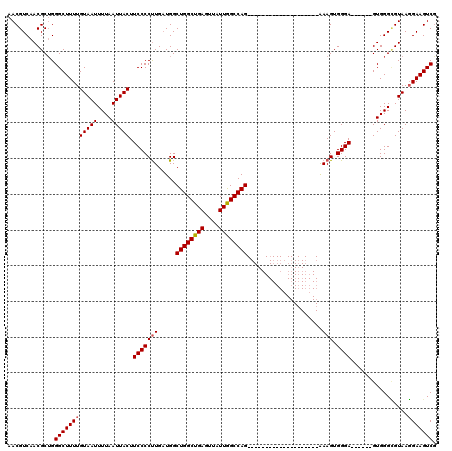
| Location | 9,674,542 – 9,674,636 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.08 |
| Mean single sequence MFE | -20.51 |
| Consensus MFE | -15.87 |
| Energy contribution | -16.11 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9674542 94 - 27905053 CGACUUCCUUACGCCCAC------UCCCACUUU--------------------CUGGCCAAUAACUCAGCCAGCCAUCAAGGGGAAGUAAUUAAAAUUACAAAAGCCCAGCGUUGACGUU .(((.((...((((....------((((.(((.--------------------(((((..........))))).....))))))).(((((....))))).........)))).)).))) ( -21.00) >DroSec_CAF1 66279 94 - 1 CGACUUCCUUACGCCCAC------UCCCACUUU--------------------CUGGCUAAUAACUCAGCCAGCCAACAAGGGGAAGUAAUUAAAAUUACAAAAGCCCUGCGUUGACGUU .(((.((...((((....------((((.(((.--------------------((((((........)))))).....))))))).(((((....))))).........)))).)).))) ( -23.10) >DroSim_CAF1 69217 94 - 1 UGACUUCCUUACGCCCAC------UCCCACUUU--------------------CUGGCCAAUAACUCAGCCAGCCAUCAAGGGGAAGUAAUUAAAAUUACAAAAGCCCAGCGUUGACGUU .(((.((...((((....------((((.(((.--------------------(((((..........))))).....))))))).(((((....))))).........)))).)).))) ( -20.90) >DroEre_CAF1 67439 98 - 1 CGACUUCAUUACUCCCAC--CCACUCCCACUUU--------------------CUGGCCAAUAACUCAGCCAGCCAUCAGGGGGAAGUAAUUAAAAUUACAAAAGCCCAGCGUUGACGUU ((((..............--...(((((.....--------------------(((((..........)))))......)))))..(((((....)))))...........))))..... ( -19.10) >DroYak_CAF1 71267 120 - 1 CGACUUCCUUACACCCACUCCCACUCCCACUUCCACUGCACCUCCCACUCCCACUGGCCAAUAACUCAGCCAGCCAUCAAGGGGAAGUAAUUAAAAUUACAAAAGCCCAGCGUUGACGUU .................................((.(((...((((.......(((((..........)))))........)))).(((((....))))).........))).))..... ( -18.46) >consensus CGACUUCCUUACGCCCAC______UCCCACUUU____________________CUGGCCAAUAACUCAGCCAGCCAUCAAGGGGAAGUAAUUAAAAUUACAAAAGCCCAGCGUUGACGUU .(((.((...((((..........((((.(((.....................(((((..........))))).....))))))).(((((....))))).........)))).)).))) (-15.87 = -16.11 + 0.24)

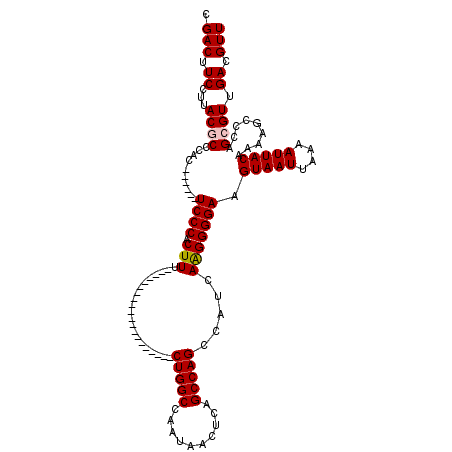
| Location | 9,674,582 – 9,674,676 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.13 |
| Mean single sequence MFE | -35.04 |
| Consensus MFE | -27.42 |
| Energy contribution | -27.38 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9674582 94 + 27905053 UUGAUGGCUGGCUGAGUUAUUGGCCAG--------------------AAAGUGGGA------GUGGGCGUAAGGAAGUCGCUGAAGGACCUGCUCCUCCAAAGUGACGGCUCGCCAGAUU .......(((((..(....)..)))))--------------------....(((((------((...(....)...((((((..((((.....))))....)))))).)))).))).... ( -31.30) >DroSec_CAF1 66319 94 + 1 UUGUUGGCUGGCUGAGUUAUUAGCCAG--------------------AAAGUGGGA------GUGGGCGUAAGGAAGUCGCUGAAGGACCUGCUCCUCCAAAGUGACGGCUCGCCAGAUU (..((..((((((((....))))))))--------------------..))..)..------...((((...((..((((((..((((.....))))....))))))..))))))..... ( -33.90) >DroSim_CAF1 69257 94 + 1 UUGAUGGCUGGCUGAGUUAUUGGCCAG--------------------AAAGUGGGA------GUGGGCGUAAGGAAGUCACUAAAGGACCUGCUCCUCCAAAGUGACGGCUCGCCAGAUU .......(((((..(....)..)))))--------------------....(((((------((...(....)...((((((..((((.....))))....)))))).)))).))).... ( -30.60) >DroEre_CAF1 67479 98 + 1 CUGAUGGCUGGCUGAGUUAUUGGCCAG--------------------AAAGUGGGAGUGG--GUGGGAGUAAUGAAGUCGCUGAAGGACCUGCUCCUCCGAAGUGACGGCUCGCCAGAUU ((.((..(((((..(....)..)))))--------------------...)).)).....--.(((((((......((((((...(((........)))..)))))).)))).))).... ( -33.40) >DroYak_CAF1 71307 120 + 1 UUGAUGGCUGGCUGAGUUAUUGGCCAGUGGGAGUGGGAGGUGCAGUGGAAGUGGGAGUGGGAGUGGGUGUAAGGAAGUCGCUGAAGGACCUGCUCCUACGAAGUGACGGCUCGCCAGAUU ....(.((((((..(....)..)))))).)...((((((.((((.(....(((((((..((..(.((((.........)))).)....))..)))))))..).)).)).))).))).... ( -46.00) >consensus UUGAUGGCUGGCUGAGUUAUUGGCCAG____________________AAAGUGGGA______GUGGGCGUAAGGAAGUCGCUGAAGGACCUGCUCCUCCAAAGUGACGGCUCGCCAGAUU .......((((((((....))))))))......................................((((...((..((((((..((((.....))))....))))))..))))))..... (-27.42 = -27.38 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:01 2006