| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,673,730 – 9,673,850 |
| Length | 120 |
| Max. P | 0.671699 |

| Location | 9,673,730 – 9,673,850 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.58 |
| Mean single sequence MFE | -42.60 |
| Consensus MFE | -30.18 |
| Energy contribution | -32.50 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.646764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9673730 120 + 27905053 GGCAAUUAAUUAAUCCUGCACUCCGUGGCCAACUUAGUUGGGCCGGCUAACUAACUAGACAGCGACUAGCAGGUUGCCGGUUGCACGUUGCAGGUUGCCGGUUGCAUAUUGCCGGGUCCA (((...........((((((...((((.(((((...)))))((((((.((((..((((.......))))..))))))))))..)))).))))))...(((((........)))))))).. ( -45.50) >DroSec_CAF1 65419 120 + 1 GGCAAUUAAUUAAUCCUGCACUCCGUGGCCAACUUAGUUGGACCGGCUAACUAACUAGACAGCGACUAGCAGGUUGCCGGUUGAACGUUGCACGUUGCCGGUUGCAUAUUGCCGGGUCCA ((((((....(((.((.(((...((((..((((...(((.(((((((.((((..((((.......))))..))))))))))).))))))))))).))).)))))...))))))....... ( -42.00) >DroSim_CAF1 68357 120 + 1 GGCAAUUAAUUAAUCCUGCACUCCGUGGCCAACUUAGUUGGACCGGCUAACUAACUAGACAGCGACUAGCAGGUUGCCGGUUGCACGUUGCAGGUUGCCGGUUGCAUAUUGCCGGGUCCA (((...........((((((...((((.(((((...)))))((((((.((((..((((.......))))..))))))))))..)))).))))))...(((((........)))))))).. ( -45.00) >DroEre_CAF1 66653 107 + 1 GGCAAUUAAUUAAUCCUGCACUCCGCGGCCAACUUAGUUGGCCCGGCUAACUAACUAGACAGCGACUAGCAGGUUGCUG-------GUUGCAGGUUGCCUAUU------UGCAGGGUCCA ............((((((((....((((((...(((((((((...))))))))).......(((((((((.....))))-------))))).)))))).....------))))))))... ( -43.90) >DroYak_CAF1 70485 120 + 1 GGCAAUUAAUUAAUCCUGCACUACGCGGCCAACUCACUUGGCCCGGCUAACUAACUAGACAGCGACUAGCAGGUUGCAGGCUUCGAGUUGCACGUUGCCUAUUGCCUAUUGCCGGGUCCA ((((((........(((((....((.((((((.....))))))))(((..((....))..))).....)))))....((((....((..((.....))))...))))))))))....... ( -36.60) >consensus GGCAAUUAAUUAAUCCUGCACUCCGUGGCCAACUUAGUUGGACCGGCUAACUAACUAGACAGCGACUAGCAGGUUGCCGGUUGCACGUUGCAGGUUGCCGGUUGCAUAUUGCCGGGUCCA (((...........((((((...((((.(((((...)))))((((((.((((..((((.......))))..))))))))))..)))).))))))...(((((........)))))))).. (-30.18 = -32.50 + 2.32)

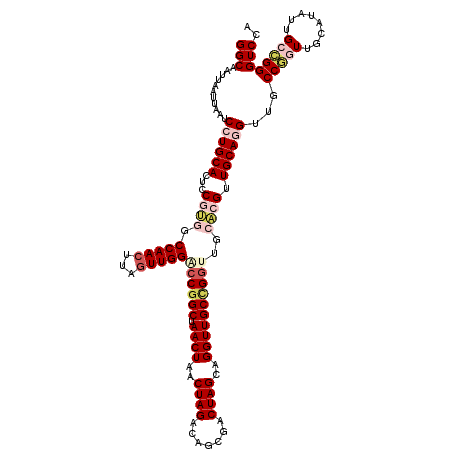
| Location | 9,673,730 – 9,673,850 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.58 |
| Mean single sequence MFE | -41.66 |
| Consensus MFE | -30.00 |
| Energy contribution | -32.96 |
| Covariance contribution | 2.96 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9673730 120 - 27905053 UGGACCCGGCAAUAUGCAACCGGCAACCUGCAACGUGCAACCGGCAACCUGCUAGUCGCUGUCUAGUUAGUUAGCCGGCCCAACUAAGUUGGCCACGGAGUGCAGGAUUAAUUAAUUGCC .......((((((.(((.....))).((((((.((((...((((((((..(((((.......)))))..))).))))).(((((...))))).))))...))))))........)))))) ( -45.80) >DroSec_CAF1 65419 120 - 1 UGGACCCGGCAAUAUGCAACCGGCAACGUGCAACGUUCAACCGGCAACCUGCUAGUCGCUGUCUAGUUAGUUAGCCGGUCCAACUAAGUUGGCCACGGAGUGCAGGAUUAAUUAAUUGCC .......((((((.(((.....))).(.((((.(((...(((((((((..(((((.......)))))..))).))))))(((((...)))))..)))...)))).)........)))))) ( -39.50) >DroSim_CAF1 68357 120 - 1 UGGACCCGGCAAUAUGCAACCGGCAACCUGCAACGUGCAACCGGCAACCUGCUAGUCGCUGUCUAGUUAGUUAGCCGGUCCAACUAAGUUGGCCACGGAGUGCAGGAUUAAUUAAUUGCC .......((((((.(((.....))).((((((.((((..(((((((((..(((((.......)))))..))).))))))(((((...))))).))))...))))))........)))))) ( -46.80) >DroEre_CAF1 66653 107 - 1 UGGACCCUGCA------AAUAGGCAACCUGCAAC-------CAGCAACCUGCUAGUCGCUGUCUAGUUAGUUAGCCGGGCCAACUAAGUUGGCCGCGGAGUGCAGGAUUAAUUAAUUGCC .....((((((------..((((((....((.((-------.(((.....))).)).)))))))).........((((((((((...))))))).)))..)))))).............. ( -37.40) >DroYak_CAF1 70485 120 - 1 UGGACCCGGCAAUAGGCAAUAGGCAACGUGCAACUCGAAGCCUGCAACCUGCUAGUCGCUGUCUAGUUAGUUAGCCGGGCCAAGUGAGUUGGCCGCGUAGUGCAGGAUUAAUUAAUUGCC .......((((((..(((...(....).))).........((((((...(((.....((((.((....)).))))..((((((.....)))))))))...))))))........)))))) ( -38.80) >consensus UGGACCCGGCAAUAUGCAACCGGCAACCUGCAACGUGCAACCGGCAACCUGCUAGUCGCUGUCUAGUUAGUUAGCCGGGCCAACUAAGUUGGCCACGGAGUGCAGGAUUAAUUAAUUGCC .......((((((..((.....))..((((((.((((...((((((((..(((((.......)))))..))).))))).(((((...))))).))))...))))))........)))))) (-30.00 = -32.96 + 2.96)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:51 2006