
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,673,398 – 9,673,613 |
| Length | 215 |
| Max. P | 0.986891 |

| Location | 9,673,398 – 9,673,511 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 95.36 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -32.08 |
| Energy contribution | -31.87 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9673398 113 - 27905053 AAUGAUGCUGCAGUUGCAGCGUUGUCAAAUGACUUGCGUAAAUUUAUCGUAAAUGCCAUCAGCUGCAGUUGAGGCUGAGCUAAGCUCCUCC--GUAUGUUCAUCCCUCCGUCAUU ((((((((((((((((..((((((((....))).((((.........)))))))))...)))))))))..((((.(((((...((......--))..)))))..))))))))))) ( -36.60) >DroSec_CAF1 65085 115 - 1 AAUGAUGCUGCAGUUGCAGCGUUGUCAAAUGACUUGCGUAAAUUUAUCGUAAAUGCCAUCAGCUGUAGUUGAGGCUGAGCUGAGCUCCUCCACGUACGUUCGUCCCUCCGUCAUU .(..(((((((....)))))))..).....(((..(((((..............(((.(((((....)))))))).((((...)))).......)))))..)))........... ( -33.80) >DroSim_CAF1 68023 115 - 1 AAUGAUUCUGCAGUUGCAGCGUUGUCAAAUGACUUGCGUAAAUUUAUCGUAAAUGCCAUCAGCUACAGUUGAGGCUGAGCUGAGCUCCUCCACGUACGUUCGUCCCUCCGUCAUU ..((((.((((....))))....))))(((((((((((.........)))))..((..((((((.((((....))))))))))))......(((......)))......)))))) ( -28.60) >consensus AAUGAUGCUGCAGUUGCAGCGUUGUCAAAUGACUUGCGUAAAUUUAUCGUAAAUGCCAUCAGCUGCAGUUGAGGCUGAGCUGAGCUCCUCCACGUACGUUCGUCCCUCCGUCAUU ((((((((((((((((..((((((((....))).((((.........)))))))))...)))))))))..((((.(((((.(.((........)).))))))..))))))))))) (-32.08 = -31.87 + -0.22)



| Location | 9,673,431 – 9,673,551 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.83 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -23.38 |
| Energy contribution | -24.02 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9673431 120 + 27905053 GCUCAGCCUCAACUGCAGCUGAUGGCAUUUACGAUAAAUUUACGCAAGUCAUUUGACAACGCUGCAACUGCAGCAUCAUUAUGCAGGCGACUGCCAGGCUCAAAUAUGCACAAAUAUGCA (((((((..(....)..)))))((((((.............((....)).(((((((...(((((....)))))........((((....))))...).)))))))))).)).....)). ( -31.90) >DroSec_CAF1 65120 120 + 1 GCUCAGCCUCAACUACAGCUGAUGGCAUUUACGAUAAAUUUACGCAAGUCAUUUGACAACGCUGCAACUGCAGCAUCAUUAUGCAGCCGACUGCCAGGCUCAAAUAUGCACAAAUAUGCA (((((((..........)))))((((((...............(((.(((....)))...(((((....))))).......)))((((........)))).....)))).)).....)). ( -31.10) >DroSim_CAF1 68058 120 + 1 GCUCAGCCUCAACUGUAGCUGAUGGCAUUUACGAUAAAUUUACGCAAGUCAUUUGACAACGCUGCAACUGCAGAAUCAUUAUGCAGCCGACUGCCAGGCUCAAAUAUGCACAAAUAUGCA (((((((..(....)..)))))((((((...............(((.(((....)))....((((....))))........)))((((........)))).....)))).)).....)). ( -28.90) >DroEre_CAF1 66347 106 + 1 --------------GCAGCUGAUGGCACUUACGAUAAAUUUACGCAAGUCAUUUGACAACGCUGCAACUGCAGCAUCAUUAUGCAGCCGACGGCCAGGCUCAAAAAUGCACAAAUAUGCA --------------(((((((.((((.(....)..........(((.(((....)))...(((((....))))).......))).)))).))))...((........)).......))). ( -29.70) >DroYak_CAF1 70180 120 + 1 GCUCAGCCUCAACUGCAGCUGAUGGCAUUUACGAUAAAUUUACGCAAGUCAUUUGACAACGCUGCAACUGCAGCAUCAUUAUGCAGCCGACGGCCAGGCUCAAAUAUGCACAAAUAUGCA (((..(((((..(((((..((((((.......(.(((((..((....)).))))).)...(((((....))))).)))))))))))..)).)))..))).......((((......)))) ( -33.90) >consensus GCUCAGCCUCAACUGCAGCUGAUGGCAUUUACGAUAAAUUUACGCAAGUCAUUUGACAACGCUGCAACUGCAGCAUCAUUAUGCAGCCGACUGCCAGGCUCAAAUAUGCACAAAUAUGCA ..............(((.......((((...............(((.(((....)))...(((((....))))).......)))((((........)))).....)))).......))). (-23.38 = -24.02 + 0.64)



| Location | 9,673,431 – 9,673,551 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.83 |
| Mean single sequence MFE | -38.94 |
| Consensus MFE | -31.48 |
| Energy contribution | -33.48 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9673431 120 - 27905053 UGCAUAUUUGUGCAUAUUUGAGCCUGGCAGUCGCCUGCAUAAUGAUGCUGCAGUUGCAGCGUUGUCAAAUGACUUGCGUAAAUUUAUCGUAAAUGCCAUCAGCUGCAGUUGAGGCUGAGC (((((....)))))...((.(((((.((((....))))........((((((((((..((((((((....))).((((.........)))))))))...))))))))))..))))).)). ( -40.30) >DroSec_CAF1 65120 120 - 1 UGCAUAUUUGUGCAUAUUUGAGCCUGGCAGUCGGCUGCAUAAUGAUGCUGCAGUUGCAGCGUUGUCAAAUGACUUGCGUAAAUUUAUCGUAAAUGCCAUCAGCUGUAGUUGAGGCUGAGC (((((....)))))...((.(((((((((((((..((....(..(((((((....)))))))..)))..))))(((((.........))))).)))))(((((....))))))))).)). ( -41.00) >DroSim_CAF1 68058 120 - 1 UGCAUAUUUGUGCAUAUUUGAGCCUGGCAGUCGGCUGCAUAAUGAUUCUGCAGUUGCAGCGUUGUCAAAUGACUUGCGUAAAUUUAUCGUAAAUGCCAUCAGCUACAGUUGAGGCUGAGC (((((....)))))...((.((((((((((.((((((((.........))))))))).((((.(((....)))..))))..............)))))(((((....))))))))).)). ( -38.40) >DroEre_CAF1 66347 106 - 1 UGCAUAUUUGUGCAUUUUUGAGCCUGGCCGUCGGCUGCAUAAUGAUGCUGCAGUUGCAGCGUUGUCAAAUGACUUGCGUAAAUUUAUCGUAAGUGCCAUCAGCUGC-------------- (((((....)))))...........(((.(..(((..(((.(..(((((((....)))))))..)...)))(((((((.........))))))))))..).)))..-------------- ( -32.50) >DroYak_CAF1 70180 120 - 1 UGCAUAUUUGUGCAUAUUUGAGCCUGGCCGUCGGCUGCAUAAUGAUGCUGCAGUUGCAGCGUUGUCAAAUGACUUGCGUAAAUUUAUCGUAAAUGCCAUCAGCUGCAGUUGAGGCUGAGC (((((....))))).......((..((((.(((((((((...(((((((((....)))))...(((....)))(((((.........))))).....))))..)))))))))))))..)) ( -42.50) >consensus UGCAUAUUUGUGCAUAUUUGAGCCUGGCAGUCGGCUGCAUAAUGAUGCUGCAGUUGCAGCGUUGUCAAAUGACUUGCGUAAAUUUAUCGUAAAUGCCAUCAGCUGCAGUUGAGGCUGAGC (((((....)))))......((((((((.((((..((....(..(((((((....)))))))..)))..))))(((((.........)))))..))))(((((....))))))))).... (-31.48 = -33.48 + 2.00)



| Location | 9,673,471 – 9,673,591 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -29.06 |
| Energy contribution | -29.66 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9673471 120 + 27905053 UACGCAAGUCAUUUGACAACGCUGCAACUGCAGCAUCAUUAUGCAGGCGACUGCCAGGCUCAAAUAUGCACAAAUAUGCAAAGCAGCUACUGAUUAUUGUCGAAAGUUGUUAGCAGAAGA ..(((..(((....)))...(((((....)))))............))).((((............((((......)))).(((((((..((((....))))..))))))).)))).... ( -33.60) >DroSec_CAF1 65160 120 + 1 UACGCAAGUCAUUUGACAACGCUGCAACUGCAGCAUCAUUAUGCAGCCGACUGCCAGGCUCAAAUAUGCACAAAUAUGCAAAGCAGCUACUGAUUAUUGUCGAAAGUUGUUAGCAGAAGA ...(((.(((....)))...(((((....))))).......((.((((........))))))....))).......(((..(((((((..((((....))))..))))))).)))..... ( -32.80) >DroSim_CAF1 68098 120 + 1 UACGCAAGUCAUUUGACAACGCUGCAACUGCAGAAUCAUUAUGCAGCCGACUGCCAGGCUCAAAUAUGCACAAAUAUGCAAAGCAGCUACUGAUUAUUGUCGAAAGUUGUUAGCAGAAGA .((....))...(((((((((((((...((((.........))))(((........))).......((((......))))..)))))...((((....))))...))))))))....... ( -29.90) >DroEre_CAF1 66373 120 + 1 UACGCAAGUCAUUUGACAACGCUGCAACUGCAGCAUCAUUAUGCAGCCGACGGCCAGGCUCAAAAAUGCACAAAUAUGCAAAGCAGCUACUGAUGAUUGUCGAAUGUUGUUAGCAGAAGA ...((....((((((((((.(((((....))))).((((((.((((((........))))......((((......)))).....))...))))))))))))))))......))...... ( -36.40) >DroYak_CAF1 70220 120 + 1 UACGCAAGUCAUUUGACAACGCUGCAACUGCAGCAUCAUUAUGCAGCCGACGGCCAGGCUCAAAUAUGCACAAAUAUGCAAAGCAGCUACUGAUGAUUGUCGAAAGUUGUUAGCAGAAGA .((....))...(((((((((((((....)))))(((((((.((((((........))))......((((......)))).....))...)))))))........))))))))....... ( -33.70) >consensus UACGCAAGUCAUUUGACAACGCUGCAACUGCAGCAUCAUUAUGCAGCCGACUGCCAGGCUCAAAUAUGCACAAAUAUGCAAAGCAGCUACUGAUUAUUGUCGAAAGUUGUUAGCAGAAGA ...(((.(((....)))...(((((....))))).......((.((((........))))))....))).......(((..(((((((..((((....))))..))))))).)))..... (-29.06 = -29.66 + 0.60)



| Location | 9,673,471 – 9,673,591 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -33.41 |
| Consensus MFE | -31.17 |
| Energy contribution | -31.57 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9673471 120 - 27905053 UCUUCUGCUAACAACUUUCGACAAUAAUCAGUAGCUGCUUUGCAUAUUUGUGCAUAUUUGAGCCUGGCAGUCGCCUGCAUAAUGAUGCUGCAGUUGCAGCGUUGUCAAAUGACUUGCGUA ..............................((((((((...((((..(((((((...........(((....))))))))))..)))).)))))))).((((.(((....)))..)))). ( -33.70) >DroSec_CAF1 65160 120 - 1 UCUUCUGCUAACAACUUUCGACAAUAAUCAGUAGCUGCUUUGCAUAUUUGUGCAUAUUUGAGCCUGGCAGUCGGCUGCAUAAUGAUGCUGCAGUUGCAGCGUUGUCAAAUGACUUGCGUA ..............................((((((((...((((..(((((((.......(((........))))))))))..)))).)))))))).((((.(((....)))..)))). ( -34.01) >DroSim_CAF1 68098 120 - 1 UCUUCUGCUAACAACUUUCGACAAUAAUCAGUAGCUGCUUUGCAUAUUUGUGCAUAUUUGAGCCUGGCAGUCGGCUGCAUAAUGAUUCUGCAGUUGCAGCGUUGUCAAAUGACUUGCGUA ..............................((((((((..(((((....)))))((((((((((........)))).)).)))).....)))))))).((((.(((....)))..)))). ( -29.60) >DroEre_CAF1 66373 120 - 1 UCUUCUGCUAACAACAUUCGACAAUCAUCAGUAGCUGCUUUGCAUAUUUGUGCAUUUUUGAGCCUGGCCGUCGGCUGCAUAAUGAUGCUGCAGUUGCAGCGUUGUCAAAUGACUUGCGUA ......((......((((.((((((.....((((((((...((((..(((((((.......(((........))))))))))..)))).))))))))...)))))).))))....))... ( -35.71) >DroYak_CAF1 70220 120 - 1 UCUUCUGCUAACAACUUUCGACAAUCAUCAGUAGCUGCUUUGCAUAUUUGUGCAUAUUUGAGCCUGGCCGUCGGCUGCAUAAUGAUGCUGCAGUUGCAGCGUUGUCAAAUGACUUGCGUA ..............................((((((((...((((..(((((((.......(((........))))))))))..)))).)))))))).((((.(((....)))..)))). ( -34.01) >consensus UCUUCUGCUAACAACUUUCGACAAUAAUCAGUAGCUGCUUUGCAUAUUUGUGCAUAUUUGAGCCUGGCAGUCGGCUGCAUAAUGAUGCUGCAGUUGCAGCGUUGUCAAAUGACUUGCGUA ..............................((((((((...((((..(((((((.......(((........))))))))))..)))).)))))))).((((.(((....)))..)))). (-31.17 = -31.57 + 0.40)

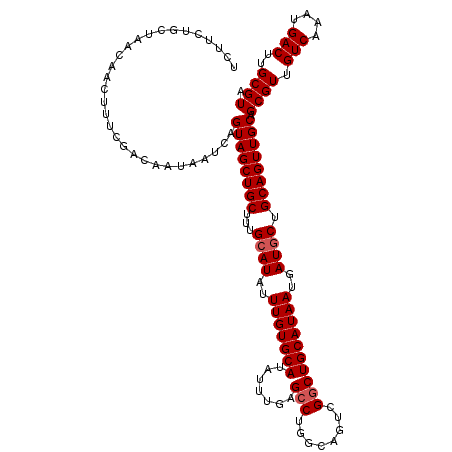
| Location | 9,673,511 – 9,673,613 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 97.39 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -24.12 |
| Energy contribution | -23.90 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9673511 102 + 27905053 AUGCAGGCGACUGCCAGGCUCAAAUAUGCACAAAUAUGCAAAGCAGCUACUGAUUAUUGUCGAAAGUUGUUAGCAGAAGAGCAAAAGCAACAGUAAGAGCAA ..((((....))))...((((.....((((......)))).(((((((..((((....))))..))))))).((......))..............)))).. ( -26.20) >DroSec_CAF1 65200 102 + 1 AUGCAGCCGACUGCCAGGCUCAAAUAUGCACAAAUAUGCAAAGCAGCUACUGAUUAUUGUCGAAAGUUGUUAGCAGAAGAGCAAAUGCAACAGCAACAGCAA .(((.(((..((((............((((......)))).(((((((..((((....))))..))))))).))))..).))...(((....)))...))). ( -26.00) >DroSim_CAF1 68138 102 + 1 AUGCAGCCGACUGCCAGGCUCAAAUAUGCACAAAUAUGCAAAGCAGCUACUGAUUAUUGUCGAAAGUUGUUAGCAGAAGAGCAAAUGCAACAGCAACAGCAA .(((.(((..((((............((((......)))).(((((((..((((....))))..))))))).))))..).))...(((....)))...))). ( -26.00) >consensus AUGCAGCCGACUGCCAGGCUCAAAUAUGCACAAAUAUGCAAAGCAGCUACUGAUUAUUGUCGAAAGUUGUUAGCAGAAGAGCAAAUGCAACAGCAACAGCAA ..((((....))))...((((.....((((......)))).(((((((..((((....))))..))))))).......))))..........((....)).. (-24.12 = -23.90 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:49 2006