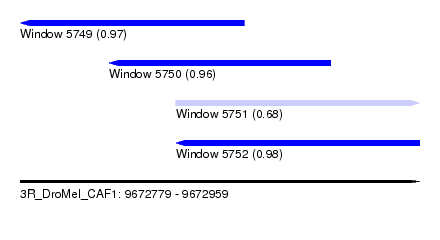
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,672,779 – 9,672,959 |
| Length | 180 |
| Max. P | 0.981239 |

| Location | 9,672,779 – 9,672,880 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.83 |
| Mean single sequence MFE | -36.72 |
| Consensus MFE | -30.01 |
| Energy contribution | -30.01 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9672779 101 - 27905053 GCCAAGAGUGAUUGCUGAGGUGGCAA---------UUGGCGCUGCCACAAAGGUGUUGCUCAUGCUCGG----------AUCUGAGUAUCUGGGAAAUGCAAAUCCAGUUAAUCACUGAA ......(((((((((((..((((((.---------.......))))))....(((((.((((((((((.----------...))))))..)))).))))).....))).))))))))... ( -33.70) >DroSec_CAF1 64465 101 - 1 GCCAAGAGUGAUUGCUGAGGUGGCAA---------UUGGCGCUGCCACAAAGGCGUUGCUCAUGCUCGG----------AUCUGAGUGUCUGGGAAAUGCAAAUCCAGUUAAUCACUGAA ......(((((((((((..((((((.---------.......))))))....(((((.((((((((((.----------...))))))..)))).))))).....))).))))))))... ( -35.00) >DroSim_CAF1 67396 101 - 1 GCCAAGAGUGAUUGCUGAGGUGGCAA---------UUGGCGCUGCCACAAAGGCGUUGCUCAUGCUCGG----------AUCUGAGUGUCUGGGAAAUGCAAAUCCAGUUAAUCACUGAA ......(((((((((((..((((((.---------.......))))))....(((((.((((((((((.----------...))))))..)))).))))).....))).))))))))... ( -35.00) >DroEre_CAF1 65650 118 - 1 GCCAAGAGUGAUUGCCGAGGUGGCAAUGGAGCUGAUUGGAGCUGCCACAAAGGUGUUGCUAAUGCU--GGUAUCUGGGUAUCUGGGUAUCUGGGAAAUGCAAAUCCAGUUGAUCACUGAA ......((((((((((...((((((.....(((......)))))))))...))).((((...(.((--((((((..(....)..)))))).)).)...))))........)))))))... ( -35.90) >DroYak_CAF1 69484 111 - 1 GCCAAGAGUGAUUGCUGAGGUGGCAA---------UUGGAGCUGCCACAAAGGUGUUGCUAAUGCUCGGGUAUCUGGCUAUCCGGGUAUCUGGGAAAUGCAAAUCCAGUUAAUCACUGAA ......(((((((((((..((((((.---------.......))))))...(((.((((.....(((((((((((((....)))))))))))))....)))))))))).))))))))... ( -44.00) >consensus GCCAAGAGUGAUUGCUGAGGUGGCAA_________UUGGCGCUGCCACAAAGGUGUUGCUCAUGCUCGG__________AUCUGAGUAUCUGGGAAAUGCAAAUCCAGUUAAUCACUGAA ......(((((((((((..((((((.................))))))....(((((.(((((((((((............))))))))..))).))))).....))).))))))))... (-30.01 = -30.01 + -0.00)


| Location | 9,672,819 – 9,672,919 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.33 |
| Mean single sequence MFE | -36.46 |
| Consensus MFE | -31.83 |
| Energy contribution | -32.23 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9672819 100 - 27905053 UAUGUCGUUUGACACAUUUAAUUGG-AGUCAAUUGGCCGGGCCAAGAGUGAUUGCUGAGGUGGCAA---------UUGGCGCUGCCACAAAGGUGUUGCUCAUGCUCGG----------A ...((((.(((((.((......)).-.))))).))))(((((...(((..((.(((...((((((.---------.......))))))...)))))..)))..))))).----------. ( -35.60) >DroSec_CAF1 64505 100 - 1 UAUGUCGUUUGACACAUUUAAUUGG-AGUCAAUUGGCCGGGCCAAGAGUGAUUGCUGAGGUGGCAA---------UUGGCGCUGCCACAAAGGCGUUGCUCAUGCUCGG----------A ...((((.(((((.((......)).-.))))).))))(((((...(((..((.(((...((((((.---------.......))))))...)))))..)))..))))).----------. ( -38.20) >DroSim_CAF1 67436 100 - 1 UAUGUCGUUUGACACAUUUAAUUGG-AGUCAAUUGGCCGGGCCAAGAGUGAUUGCUGAGGUGGCAA---------UUGGCGCUGCCACAAAGGCGUUGCUCAUGCUCGG----------A ...((((.(((((.((......)).-.))))).))))(((((...(((..((.(((...((((((.---------.......))))))...)))))..)))..))))).----------. ( -38.20) >DroEre_CAF1 65690 118 - 1 CAUGUCGUUUGACACAUUUAAUUGGAAGUCAAUUGGCCGGGCCAAGAGUGAUUGCCGAGGUGGCAAUGGAGCUGAUUGGAGCUGCCACAAAGGUGUUGCUAAUGCU--GGUAUCUGGGUA ....(((.(((((.((......))...))))).)))((((((((..((..((.(((...((((((.....(((......)))))))))...)))))..)).....)--)))..))))... ( -32.80) >DroYak_CAF1 69524 111 - 1 UAUGUCGCUUGACACAUUUAAUUGGAAGUCAAUUGGCCGGGCCAAGAGUGAUUGCUGAGGUGGCAA---------UUGGAGCUGCCACAAAGGUGUUGCUAAUGCUCGGGUAUCUGGCUA ..((((....))))............(((((..((.((((((....((..((.(((...((((((.---------.......))))))...)))))..))...)))))).))..))))). ( -37.50) >consensus UAUGUCGUUUGACACAUUUAAUUGG_AGUCAAUUGGCCGGGCCAAGAGUGAUUGCUGAGGUGGCAA_________UUGGCGCUGCCACAAAGGUGUUGCUCAUGCUCGG__________A ...((((.(((((.((......))...))))).))))(((((...(((..((.(((...((((((.................))))))...)))))..)))..)))))............ (-31.83 = -32.23 + 0.40)

| Location | 9,672,849 – 9,672,959 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.53 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -22.26 |
| Energy contribution | -22.10 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9672849 110 + 27905053 GCCAA---------UUGCCACCUCAGCAAUCACUCUUGGCCCGGCCAAUUGACU-CCAAUUAAAUGUGUCAAACGACAUAGACCGUUAACACGCCACGUCUGGCAUUGAGACCACUAGCU ((..(---------((((.......)))))..(((...(((.(((.(((((...-.)))))....((((..((((........)))).)))).....))).)))...))).......)). ( -23.60) >DroSec_CAF1 64535 110 + 1 GCCAA---------UUGCCACCUCAGCAAUCACUCUUGGCCCGGCCAAUUGACU-CCAAUUAAAUGUGUCAAACGACAUAGACCGUUAACACGCCACGUCUGGCAUUGAGACCACUAGCU ((..(---------((((.......)))))..(((...(((.(((.(((((...-.)))))....((((..((((........)))).)))).....))).)))...))).......)). ( -23.60) >DroSim_CAF1 67466 110 + 1 GCCAA---------UUGCCACCUCAGCAAUCACUCUUGGCCCGGCCAAUUGACU-CCAAUUAAAUGUGUCAAACGACAUAGACCGUUAACACGCCACGUCUGGCAUUGAGACCACUAGCU ((..(---------((((.......)))))..(((...(((.(((.(((((...-.)))))....((((..((((........)))).)))).....))).)))...))).......)). ( -23.60) >DroEre_CAF1 65728 120 + 1 UCCAAUCAGCUCCAUUGCCACCUCGGCAAUCACUCUUGGCCCGGCCAAUUGACUUCCAAUUAAAUGUGUCAAACGACAUGGACCGUUAACACGCCAAGUCUGGCAUUGAGACCACUAGCU .......((((..((((((.....))))))...(((..(..((((((((((.....)))))....(((((....))))))).))).......((((....)))).)..))).....)))) ( -30.50) >DroYak_CAF1 69564 111 + 1 UCCAA---------UUGCCACCUCAGCAAUCACUCUUGGCCCGGCCAAUUGACUUCCAAUUAAAUGUGUCAAGCGACAUAGGCCGUUAACACGCCAAGUCGGGCAUUGAGACCACUAGCU ....(---------((((.......)))))..(((...(((((((.(((((.....)))))...((((((....))))))(((.((....)))))..)))))))...))).......... ( -31.70) >consensus GCCAA_________UUGCCACCUCAGCAAUCACUCUUGGCCCGGCCAAUUGACU_CCAAUUAAAUGUGUCAAACGACAUAGACCGUUAACACGCCACGUCUGGCAUUGAGACCACUAGCU ..............((((.......))))...(((...(((.(((.(((((.....)))))....((((..((((........)))).)))).....))).)))...))).......... (-22.26 = -22.10 + -0.16)

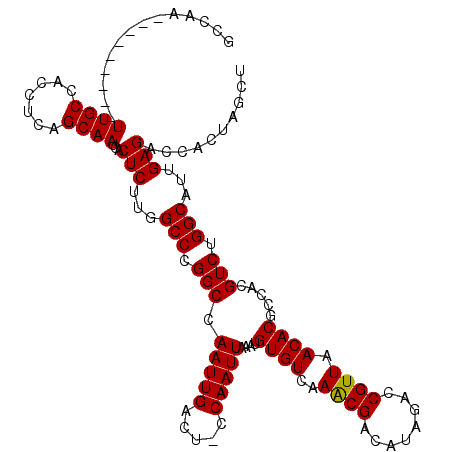
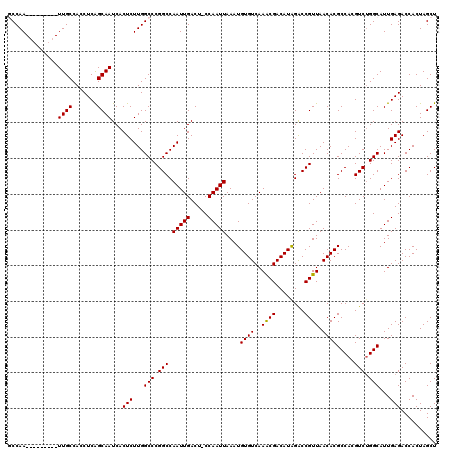
| Location | 9,672,849 – 9,672,959 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.53 |
| Mean single sequence MFE | -39.34 |
| Consensus MFE | -33.18 |
| Energy contribution | -33.46 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9672849 110 - 27905053 AGCUAGUGGUCUCAAUGCCAGACGUGGCGUGUUAACGGUCUAUGUCGUUUGACACAUUUAAUUGG-AGUCAAUUGGCCGGGCCAAGAGUGAUUGCUGAGGUGGCAA---------UUGGC .(((..((((((....((((((((((((((....)))..)))))))..(((((.((......)).-.))))).)))).))))))..)))(((((((.....)))))---------))... ( -38.10) >DroSec_CAF1 64535 110 - 1 AGCUAGUGGUCUCAAUGCCAGACGUGGCGUGUUAACGGUCUAUGUCGUUUGACACAUUUAAUUGG-AGUCAAUUGGCCGGGCCAAGAGUGAUUGCUGAGGUGGCAA---------UUGGC .(((..((((((....((((((((((((((....)))..)))))))..(((((.((......)).-.))))).)))).))))))..)))(((((((.....)))))---------))... ( -38.10) >DroSim_CAF1 67466 110 - 1 AGCUAGUGGUCUCAAUGCCAGACGUGGCGUGUUAACGGUCUAUGUCGUUUGACACAUUUAAUUGG-AGUCAAUUGGCCGGGCCAAGAGUGAUUGCUGAGGUGGCAA---------UUGGC .(((..((((((....((((((((((((((....)))..)))))))..(((((.((......)).-.))))).)))).))))))..)))(((((((.....)))))---------))... ( -38.10) >DroEre_CAF1 65728 120 - 1 AGCUAGUGGUCUCAAUGCCAGACUUGGCGUGUUAACGGUCCAUGUCGUUUGACACAUUUAAUUGGAAGUCAAUUGGCCGGGCCAAGAGUGAUUGCCGAGGUGGCAAUGGAGCUGAUUGGA ..((((((((..((.(((((..((((((((((((((((......))).)))))))...........(((((.(((((...)))))...))))))))))).))))).))..))).))))). ( -45.70) >DroYak_CAF1 69564 111 - 1 AGCUAGUGGUCUCAAUGCCCGACUUGGCGUGUUAACGGCCUAUGUCGCUUGACACAUUUAAUUGGAAGUCAAUUGGCCGGGCCAAGAGUGAUUGCUGAGGUGGCAA---------UUGGA .(((..(((((...(((((......))))).....(((((..((((....)))).....(((((.....)))))))))))))))..)))(((((((.....)))))---------))... ( -36.70) >consensus AGCUAGUGGUCUCAAUGCCAGACGUGGCGUGUUAACGGUCUAUGUCGUUUGACACAUUUAAUUGG_AGUCAAUUGGCCGGGCCAAGAGUGAUUGCUGAGGUGGCAA_________UUGGC .(((..((((((....((((((((((((((....)))..)))))))..(((((.((......))...))))).)))).))))))..)))..(((((.....))))).............. (-33.18 = -33.46 + 0.28)

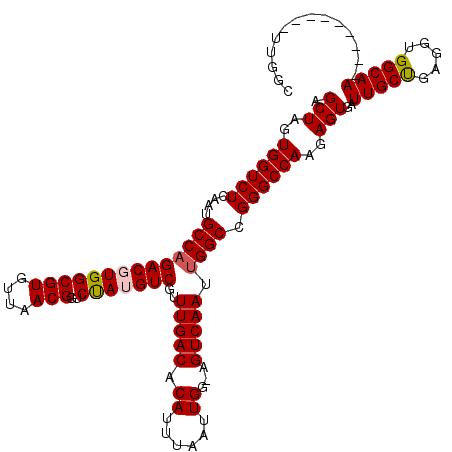
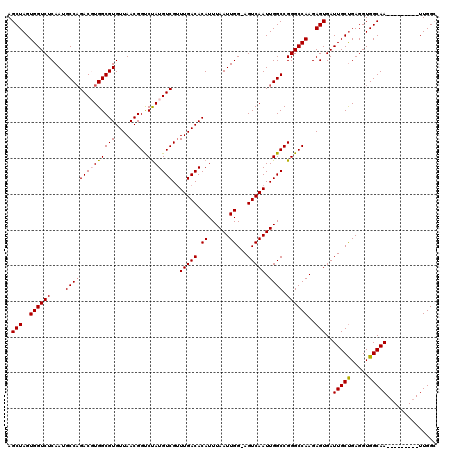
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:43 2006