
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,670,823 – 9,671,004 |
| Length | 181 |
| Max. P | 0.999954 |

| Location | 9,670,823 – 9,670,942 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.24 |
| Mean single sequence MFE | -38.16 |
| Consensus MFE | -31.50 |
| Energy contribution | -31.58 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9670823 119 - 27905053 AACGGCCACACAAUCGCCAUAAGUUUUGGCCGCUCUGCAAUUAUUUGCGGAAUUCCUUUUGGAAUAUAUAUUGACUCGGACUGGAGUGCUCGGC-CAGACUGGUCAACUCAACCAGGAGA ...((((........((((.......)))).(((((((((....)))))))(((((....)))))........((((......))))))..)))-)...(((((.......))))).... ( -37.70) >DroSec_CAF1 62623 119 - 1 AACGGCCACACAAUCGCCAUAAGUUUUGGCCGCUCUGCAAUUAUUUGCGGAAUUCCUUUUGGAAUAUAUAUUGACUCGGACUGGAGUGCCCGGC-CAGACUGGCCAACUCAACCAGGAGA ...(((((.......((.....))((((((((.(((((((....)))))))(((((....)))))........((((......))))...))))-)))).)))))..(((......))). ( -38.10) >DroSim_CAF1 65534 119 - 1 AACGGCCACACAAUCGCCAUAAGUUUUGGCCGCUCUGCAAUUAUUUGCGGAAUUCCUUUUGGAAUAUAUAUUGACUCGGACUGAAGUGCCCGGC-CAGACUGGCCAACUCAACCAGGAGA ...(((((.......((.....))((((((((.(((((((....)))))))(((((....)))))............((((....)).))))))-)))).)))))..(((......))). ( -35.90) >DroEre_CAF1 63815 120 - 1 AGCGGGCACACAAUCGCCAUAAGUUUUGGCCGCUCUGCAAUUAUUUGCGGAAUUCCUUUCGGAAUAUAUAUUGACUCGCACUGGAGUGCCAGGCCCAGACUGGCCAACUCAACCAGGAGA .(((((((.......((((.......))))...(((((((....)))))))(((((....)))))......)).))))).((((((((((((.......)))))..)))...)))).... ( -39.50) >DroYak_CAF1 67519 120 - 1 AGCGGCCACACAAUCGCCAUAAGUUUUGGCCGCUCUGCAAUUAUUUGCGGAAUUCCUUUCGGAAUAUAUAUUGACUCGGACUGGAGUGCCUGGCCCAGACUGGCCAACUCAACCAGGAGA .(((((((.((...........))..)))))))(((((((....)))))))(((((....))))).........(((((....((((...(((((......)))))))))..))..))). ( -39.60) >consensus AACGGCCACACAAUCGCCAUAAGUUUUGGCCGCUCUGCAAUUAUUUGCGGAAUUCCUUUUGGAAUAUAUAUUGACUCGGACUGGAGUGCCCGGC_CAGACUGGCCAACUCAACCAGGAGA ...(((((.......(((....((((..((((.(((((((....)))))))(((((....)))))...........))).)..))))....)))......)))))..(((......))). (-31.50 = -31.58 + 0.08)

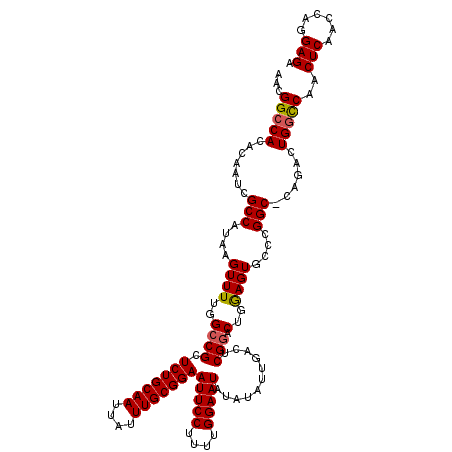

| Location | 9,670,862 – 9,670,982 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.72 |
| Mean single sequence MFE | -35.54 |
| Consensus MFE | -33.04 |
| Energy contribution | -32.84 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.83 |
| SVM RNA-class probability | 0.999954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9670862 120 + 27905053 UCCGAGUCAAUAUAUAUUCCAAAAGGAAUUCCGCAAAUAAUUGCAGAGCGGCCAAAACUUAUGGCGAUUGUGUGGCCGUUCGGCCAGGCGAUGCUCACUGAAAAAAAAAAUGUUUACACA .....((.(((((...((((....))))(((.((((....)))).((((.((((.......)))).(((((.(((((....))))).)))))))))...))).......))))).))... ( -36.40) >DroSec_CAF1 62662 120 + 1 UCCGAGUCAAUAUAUAUUCCAAAAGGAAUUCCGCAAAUAAUUGCAGAGCGGCCAAAACUUAUGGCGAUUGUGUGGCCGUUCGGCCAGGCAAAGCUCACUGAAAAAAAAAAGUUUUACAAA ......(((......(((((....)))))...((((....)))).((((.((((.......))))..((((.(((((....))))).)))).))))..)))................... ( -33.90) >DroSim_CAF1 65573 117 + 1 UCCGAGUCAAUAUAUAUUCCAAAAGGAAUUCCGCAAAUAAUUGCAGAGCGGCCAAAACUUAUGGCGAUUGUGUGGCCGUUCGGCCAGGCAAAGCUCAGUGAAA---AAAAGUUUAACAAA ......(((......(((((....)))))...((((....)))).((((.((((.......))))..((((.(((((....))))).)))).))))..)))..---.............. ( -34.00) >DroEre_CAF1 63855 113 + 1 UGCGAGUCAAUAUAUAUUCCGAAAGGAAUUCCGCAAAUAAUUGCAGAGCGGCCAAAACUUAUGGCGAUUGUGUGCCCGCUCGGCCAGGCGAGGCUCACUGAAAGAAAAC-GUA------A ((((((((.......(((((....)))))((.((((....)))).))...((((.......))))))))(((.(((((((......)))).))).)))..........)-)))------. ( -35.50) >DroYak_CAF1 67559 116 + 1 UCCGAGUCAAUAUAUAUUCCGAAAGGAAUUCCGCAAAUAAUUGCAGAGCGGCCAAAACUUAUGGCGAUUGUGUGGCCGCUCGGCUAAGCGAGGCUCACUGAAAGAAAAU-GUU-UA--AA ...(((((.......(((((....))))).((((((....)))).(((((((((..((...........)).)))))))))))........))))).............-...-..--.. ( -37.90) >consensus UCCGAGUCAAUAUAUAUUCCAAAAGGAAUUCCGCAAAUAAUUGCAGAGCGGCCAAAACUUAUGGCGAUUGUGUGGCCGUUCGGCCAGGCGAAGCUCACUGAAAAAAAAAAGUUUUACAAA ......(((......(((((....)))))...((((....)))).((((.((((.......))))..((((.(((((....))))).)))).))))..)))................... (-33.04 = -32.84 + -0.20)

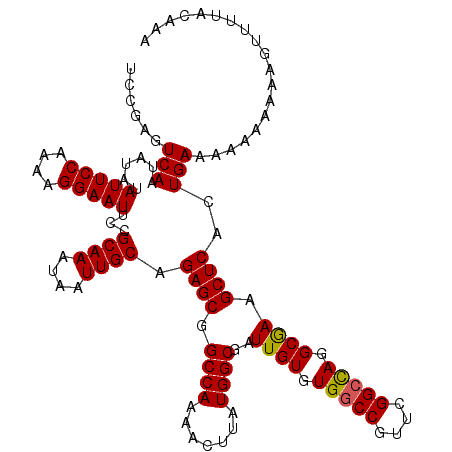

| Location | 9,670,862 – 9,670,982 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.72 |
| Mean single sequence MFE | -33.06 |
| Consensus MFE | -27.96 |
| Energy contribution | -28.56 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.990858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9670862 120 - 27905053 UGUGUAAACAUUUUUUUUUUCAGUGAGCAUCGCCUGGCCGAACGGCCACACAAUCGCCAUAAGUUUUGGCCGCUCUGCAAUUAUUUGCGGAAUUCCUUUUGGAAUAUAUAUUGACUCGGA (((....))).........((((((.((...(..(((((....)))))..)....((((.......)))).))(((((((....)))))))(((((....)))))...))))))...... ( -32.50) >DroSec_CAF1 62662 120 - 1 UUUGUAAAACUUUUUUUUUUCAGUGAGCUUUGCCUGGCCGAACGGCCACACAAUCGCCAUAAGUUUUGGCCGCUCUGCAAUUAUUUGCGGAAUUCCUUUUGGAAUAUAUAUUGACUCGGA ...................((((((.((.(((..(((((....)))))..)))..((((.......)))).))(((((((....)))))))(((((....)))))...))))))...... ( -33.10) >DroSim_CAF1 65573 117 - 1 UUUGUUAAACUUUU---UUUCACUGAGCUUUGCCUGGCCGAACGGCCACACAAUCGCCAUAAGUUUUGGCCGCUCUGCAAUUAUUUGCGGAAUUCCUUUUGGAAUAUAUAUUGACUCGGA ..............---.....(((((..(((..(((((....)))))..)))((((((.......))))...(((((((....)))))))(((((....))))).......))))))). ( -33.40) >DroEre_CAF1 63855 113 - 1 U------UAC-GUUUUCUUUCAGUGAGCCUCGCCUGGCCGAGCGGGCACACAAUCGCCAUAAGUUUUGGCCGCUCUGCAAUUAUUUGCGGAAUUCCUUUCGGAAUAUAUAUUGACUCGCA .------...-...........(((((....((..((((((((.(((........)))....).)))))))))(((((((....)))))))(((((....))))).........))))). ( -34.00) >DroYak_CAF1 67559 116 - 1 UU--UA-AAC-AUUUUCUUUCAGUGAGCCUCGCUUAGCCGAGCGGCCACACAAUCGCCAUAAGUUUUGGCCGCUCUGCAAUUAUUUGCGGAAUUCCUUUCGGAAUAUAUAUUGACUCGGA ..--..-...-............(((((...))))).(((((((((((.((...........))..)))))))(((((((....)))))))(((((....)))))..........)))). ( -32.30) >consensus UUUGUAAAACUUUUUUCUUUCAGUGAGCCUCGCCUGGCCGAACGGCCACACAAUCGCCAUAAGUUUUGGCCGCUCUGCAAUUAUUUGCGGAAUUCCUUUUGGAAUAUAUAUUGACUCGGA ...................((((((.((...(..(((((....)))))..)....((((.......)))).))(((((((....)))))))(((((....)))))...))))))...... (-27.96 = -28.56 + 0.60)


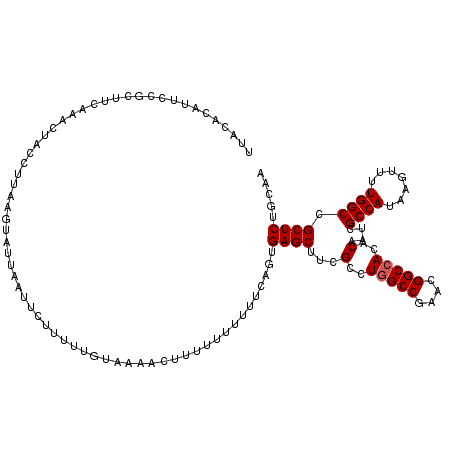
| Location | 9,670,902 – 9,671,004 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.39 |
| Mean single sequence MFE | -30.37 |
| Consensus MFE | -26.04 |
| Energy contribution | -25.60 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9670902 102 + 27905053 UUGCAGAGCGGCCAAAACUUAUGGCGAUUGUGUGGCCGUUCGGCCAGGCGAUGCUCACUGAAAAAAAAAAUGUUUACACAUA-AUUUAAUACUAAAGGUAUAU----------------- .....((((.((((.......)))).(((((.(((((....))))).))))))))).............((((....)))).-.....((((.....))))..----------------- ( -29.30) >DroSec_CAF1 62702 120 + 1 UUGCAGAGCGGCCAAAACUUAUGGCGAUUGUGUGGCCGUUCGGCCAGGCAAAGCUCACUGAAAAAAAAAAGUUUUACAAAAAGAAUUAAUACUUAAGGUAGUUUGAAGCGGAAUGUGUAA .((((((((.((((.......))))..((((.(((((....))))).)))).)))).(((....(((..((((((......))))))..(((.....))).)))....)))....)))). ( -31.70) >DroSim_CAF1 65613 117 + 1 UUGCAGAGCGGCCAAAACUUAUGGCGAUUGUGUGGCCGUUCGGCCAGGCAAAGCUCAGUGAAA---AAAAGUUUAACAAAAAGAAUUAAUACUUAAGGUAGUUUGAAGCGGAAUGUGUAA ((((.((((.((((.......))))..((((.(((((....))))).)))).)))).......---................((((((...(....).))))))...))))......... ( -30.80) >DroYak_CAF1 67599 116 + 1 UUGCAGAGCGGCCAAAACUUAUGGCGAUUGUGUGGCCGCUCGGCUAAGCGAGGCUCACUGAAAGAAAAU-GUU-UA--AACACAGUAAAUGCCUAAUGAAAUACGAUGAGAAAUGCGACG ((((((((((((((..((...........)).))))))))).........((((..((((.........-...-..--....))))....))))...................))))).. ( -29.67) >consensus UUGCAGAGCGGCCAAAACUUAUGGCGAUUGUGUGGCCGUUCGGCCAGGCAAAGCUCACUGAAAAAAAAAAGUUUUACAAAAAGAAUUAAUACUUAAGGUAGUUUGAAGCGGAAUGUGUAA .....((((.((((.......))))..((((.(((((....))))).)))).))))................................................................ (-26.04 = -25.60 + -0.44)



| Location | 9,670,902 – 9,671,004 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.39 |
| Mean single sequence MFE | -25.58 |
| Consensus MFE | -18.27 |
| Energy contribution | -18.53 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9670902 102 - 27905053 -----------------AUAUACCUUUAGUAUUAAAU-UAUGUGUAAACAUUUUUUUUUUCAGUGAGCAUCGCCUGGCCGAACGGCCACACAAUCGCCAUAAGUUUUGGCCGCUCUGCAA -----------------...........(....(((.-.((((....))))..)))....).((((((...(..(((((....)))))..)....((((.......)))).)))).)).. ( -23.80) >DroSec_CAF1 62702 120 - 1 UUACACAUUCCGCUUCAAACUACCUUAAGUAUUAAUUCUUUUUGUAAAACUUUUUUUUUUCAGUGAGCUUUGCCUGGCCGAACGGCCACACAAUCGCCAUAAGUUUUGGCCGCUCUGCAA ...........((.......(((...(((........)))...)))..................((((.(((..(((((....)))))..)))..((((.......)))).)))).)).. ( -24.10) >DroSim_CAF1 65613 117 - 1 UUACACAUUCCGCUUCAAACUACCUUAAGUAUUAAUUCUUUUUGUUAAACUUUU---UUUCACUGAGCUUUGCCUGGCCGAACGGCCACACAAUCGCCAUAAGUUUUGGCCGCUCUGCAA ...........((.............((((.(((((.......)))))))))..---.......((((.(((..(((((....)))))..)))..((((.......)))).)))).)).. ( -24.80) >DroYak_CAF1 67599 116 - 1 CGUCGCAUUUCUCAUCGUAUUUCAUUAGGCAUUUACUGUGUU--UA-AAC-AUUUUCUUUCAGUGAGCCUCGCUUAGCCGAGCGGCCACACAAUCGCCAUAAGUUUUGGCCGCUCUGCAA ....((....(((((.(.....).((((((((.....)))))--))-)..-...........)))))....))...((.(((((((((.((...........))..))))))))).)).. ( -29.60) >consensus UUACACAUUCCGCUUCAAACUACCUUAAGUAUUAAUUCUUUUUGUAAAACUUUUUUUUUUCAGUGAGCUUCGCCUGGCCGAACGGCCACACAAUCGCCAUAAGUUUUGGCCGCUCUGCAA ................................................................((((...(..(((((....)))))..)....((((.......)))).))))..... (-18.27 = -18.53 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:36 2006