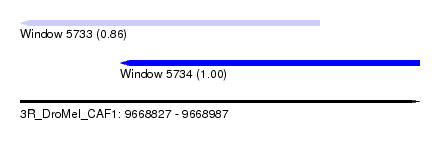
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,668,827 – 9,668,987 |
| Length | 160 |
| Max. P | 0.998817 |

| Location | 9,668,827 – 9,668,947 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.39 |
| Mean single sequence MFE | -26.21 |
| Consensus MFE | -25.42 |
| Energy contribution | -25.42 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.862921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
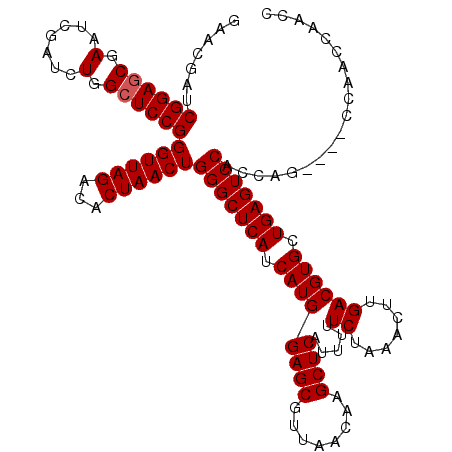
>3R_DroMel_CAF1 9668827 120 - 27905053 UAACUGGGCUCAUCAUGGAGCGUUAACAAGCUCAUUUUUCUAAACUUGACGUGCUGAGUCCACCAGCCAGCCAACCAACCAACUAGAGAUCCACUUAGGUAAAAGAUCUUCGCAGAUUAU ....((((((((.((((((((........)))).....((.......)))))).))))))))...................(((.(((.....))).)))....(((((....))))).. ( -25.90) >DroSec_CAF1 60635 116 - 1 UAACUGGGCUCAUCAUGGAGCGUUAACAAGCUCAUUUUUCUAAACUUGACGUGCUGAGUCCACCAG----CCAACCAACCAACUAGAGAUCCACUUAGGCAAAAGAUCUUCGCAGAUUAU ....((((((((.((((((((........)))).....((.......)))))).))))))))...(----((.........................)))....(((((....))))).. ( -26.51) >DroSim_CAF1 63541 116 - 1 UAACUGGGCUCAUCAUGGAGCGUUAACAAGCUCAUUUUUCUAAACUUGACGUGCUGAGUCCACCAG----CCAACCAACCAACUAGAGAUCCACUUAGGCAAAAGAUCUUCGCAGAUUAU ....((((((((.((((((((........)))).....((.......)))))).))))))))...(----((.........................)))....(((((....))))).. ( -26.51) >DroEre_CAF1 61461 112 - 1 UAACUGGGCUCAUCAUGGAGCGUUAACAAGCUCAUUUUUCUAAACUUGACGUGCUGAGUCCACCAA--------CCAUCCAACUAGAGAUCCACUUAGGCAAAAGAUCUUCGCAGAUUAU ....((((((((.((((((((........)))).....((.......)))))).))))))))....--------...........((((((.............)))))).......... ( -25.42) >DroYak_CAF1 65416 112 - 1 UAACUGGGCUCAUCAUGGAGCGUUAACAAGCUCAUUUUUCUAAACUUGACGUGCUGAGUCCACCAG--------CCAUCCAACUAGAGAUCCACUUAGGCAAAAGAUCUUCGCAGAUUAU ....((((((((.((((((((........)))).....((.......)))))).))))))))...(--------((.....................)))....(((((....))))).. ( -26.70) >consensus UAACUGGGCUCAUCAUGGAGCGUUAACAAGCUCAUUUUUCUAAACUUGACGUGCUGAGUCCACCAG____CCAACCAACCAACUAGAGAUCCACUUAGGCAAAAGAUCUUCGCAGAUUAU ....((((((((.((((((((........)))).....((.......)))))).)))))))).......................((((((.............)))))).......... (-25.42 = -25.42 + -0.00)


| Location | 9,668,867 – 9,668,987 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.39 |
| Mean single sequence MFE | -37.64 |
| Consensus MFE | -34.30 |
| Energy contribution | -34.50 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9668867 120 - 27905053 GAACGAUCGGAGCGAAUCGAUCUGGCUCCGGGUUAGACACUAACUGGGCUCAUCAUGGAGCGUUAACAAGCUCAUUUUUCUAAACUUGACGUGCUGAGUCCACCAGCCAGCCAACCAACC ....((((((......))))))(((((..(((((((...)))))((((((((.((((((((........)))).....((.......)))))).)))))))))))))))........... ( -38.20) >DroSec_CAF1 60675 116 - 1 GAACGAUCGGAGCGAAUCGAUCUGGCUCCGGGUUAGACACUAACUGGGCUCAUCAUGGAGCGUUAACAAGCUCAUUUUUCUAAACUUGACGUGCUGAGUCCACCAG----CCAACCAACC ....((((((......))))))(((((..(((((((...)))))((((((((.((((((((........)))).....((.......)))))).))))))))))))----)))....... ( -38.20) >DroSim_CAF1 63581 116 - 1 GAACGAUCGGAGCGAAUCGAUCUGGCUCCGGGUUAGACACUAACUGGGCUCAUCAUGGAGCGUUAACAAGCUCAUUUUUCUAAACUUGACGUGCUGAGUCCACCAG----CCAACCAACC ....((((((......))))))(((((..(((((((...)))))((((((((.((((((((........)))).....((.......)))))).))))))))))))----)))....... ( -38.20) >DroEre_CAF1 61501 112 - 1 GAACGAUCGGAGCGAAUCGAUCUGGCUCCGGGUUAGACACUAACUGGGCUCAUCAUGGAGCGUUAACAAGCUCAUUUUUCUAAACUUGACGUGCUGAGUCCACCAA--------CCAUCC .......((((((.(.......).))))))((((((...))))))(((((((.((((((((........)))).....((.......)))))).))))))).....--------...... ( -35.60) >DroYak_CAF1 65456 112 - 1 GAACGAUCGGACCGAAUCGAUCUGGCUCCGGGUUAGACACUAACUGGGCUCAUCAUGGAGCGUUAACAAGCUCAUUUUUCUAAACUUGACGUGCUGAGUCCACCAG--------CCAUCC ....((((((......))))))(((((..(((((((...)))))((((((((.((((((((........)))).....((.......)))))).))))))))))))--------)))... ( -38.00) >consensus GAACGAUCGGAGCGAAUCGAUCUGGCUCCGGGUUAGACACUAACUGGGCUCAUCAUGGAGCGUUAACAAGCUCAUUUUUCUAAACUUGACGUGCUGAGUCCACCAG____CCAACCAACC .......((((((.(.......).))))))((((((...))))))(((((((.((((((((........)))).....((.......)))))).)))))))................... (-34.30 = -34.50 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:27 2006