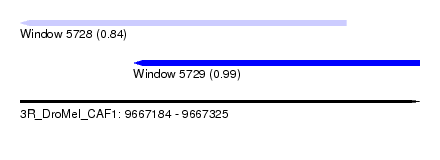
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,667,184 – 9,667,325 |
| Length | 141 |
| Max. P | 0.990207 |

| Location | 9,667,184 – 9,667,299 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.62 |
| Mean single sequence MFE | -34.94 |
| Consensus MFE | -35.18 |
| Energy contribution | -34.94 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.19 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9667184 115 - 27905053 -----AUCGUUGGCGUUGCGGUGUUUUGUUUAUUUUGAGGCUAUAAUUUAAGCGGGUAAUUGCACGCUGGCGGCCAGAAUUUGUAACUAAACUGCAAAUUAGCAGCCAAUUCCCAGGCCG -----...((..((((.(((((..((((((((..(((......)))..))))))))..)))))))))..))((((.(((((.((..((((........))))..)).)))))...)))). ( -35.10) >DroSec_CAF1 58999 115 - 1 -----AUCGUUGGCGUUGCGGUGUUUUGUUUAUUUUGAGGCUAUAAUUUAAGCGGGUAAUUGCACGCUGGCGGCCAGAAUUUGUAACUAAACUGCAAAUUAGCAGCCAAUUCCCAGGCCG -----...((..((((.(((((..((((((((..(((......)))..))))))))..)))))))))..))((((.(((((.((..((((........))))..)).)))))...)))). ( -35.10) >DroSim_CAF1 61952 115 - 1 -----AUCGUUGGCGUUGCGGUGUUUUGUUUAUUUUGAGGCUAUAAUUUAAGCGGGUAAUUGCACGCUGGCGGCCAGAAUUUGUAACUAAACUGCAAAUUAGCAGCCAAUUCCCAGGCCG -----...((..((((.(((((..((((((((..(((......)))..))))))))..)))))))))..))((((.(((((.((..((((........))))..)).)))))...)))). ( -35.10) >DroEre_CAF1 59779 120 - 1 UGAUUCUCGUUGGCGUUGCGGUGUUUUGUUUAUUUUGAGGCUAUAAUUUAGGCGGGUAAUUGCACGCUGGCGGCCAGAAUUUGUAACUAAACUGCAAAUUAGCAGCCAAUUCCCAGGCCG ........((..((((.(((((..((((((((..(((......)))..))))))))..)))))))))..))((((.(((((.((..((((........))))..)).)))))...)))). ( -34.70) >DroYak_CAF1 63694 119 - 1 -GAUUCUCGUUGGCGUUGCGGUGUUUUGUUUAUUUUGAGGCUAUAAUUUAGGCGGGUAAUUGCACGCUGGCGGCCAGAAUUUGUAACUAAACUGCAAAUUAGCAGCCAAUUCCCAGGCCG -.......((..((((.(((((..((((((((..(((......)))..))))))))..)))))))))..))((((.(((((.((..((((........))))..)).)))))...)))). ( -34.70) >consensus _____AUCGUUGGCGUUGCGGUGUUUUGUUUAUUUUGAGGCUAUAAUUUAAGCGGGUAAUUGCACGCUGGCGGCCAGAAUUUGUAACUAAACUGCAAAUUAGCAGCCAAUUCCCAGGCCG ........((..((((.(((((..((((((((..(((......)))..))))))))..)))))))))..))((((.(((((.((..((((........))))..)).)))))...)))). (-35.18 = -34.94 + -0.24)



| Location | 9,667,224 – 9,667,325 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.95 |
| Mean single sequence MFE | -35.82 |
| Consensus MFE | -32.06 |
| Energy contribution | -31.66 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9667224 101 - 27905053 UCUGGAGUUAAGCGAACUGACCAUC-----C--------------AUCGUUGGCGUUGCGGUGUUUUGUUUAUUUUGAGGCUAUAAUUUAAGCGGGUAAUUGCACGCUGGCGGCCAGAAU (((((((((.....)))).......-----.--------------.((((..((((.(((((..((((((((..(((......)))..))))))))..)))))))))..))))))))).. ( -31.90) >DroSec_CAF1 59039 101 - 1 UCUGGAGUUAAGUGAACUGACCAUC-----C--------------AUCGUUGGCGUUGCGGUGUUUUGUUUAUUUUGAGGCUAUAAUUUAAGCGGGUAAUUGCACGCUGGCGGCCAGAAU (((((.((((.(....)))))....-----.--------------.((((..((((.(((((..((((((((..(((......)))..))))))))..)))))))))..))))))))).. ( -32.70) >DroSim_CAF1 61992 101 - 1 UCUGGAGUUAAGUGAACUGACCAUC-----C--------------AUCGUUGGCGUUGCGGUGUUUUGUUUAUUUUGAGGCUAUAAUUUAAGCGGGUAAUUGCACGCUGGCGGCCAGAAU (((((.((((.(....)))))....-----.--------------.((((..((((.(((((..((((((((..(((......)))..))))))))..)))))))))..))))))))).. ( -32.70) >DroEre_CAF1 59819 120 - 1 UCUGGAGUUAAGUGAGCUGACCGUCGCGUCCAGCUCUCGUUGAUUCUCGUUGGCGUUGCGGUGUUUUGUUUAUUUUGAGGCUAUAAUUUAGGCGGGUAAUUGCACGCUGGCGGCCAGAAU ((((((((((((.((((((..((...))..)))))).).)))))))((((..((((.(((((..((((((((..(((......)))..))))))))..)))))))))..)))).)))).. ( -43.70) >DroYak_CAF1 63734 113 - 1 UCUGGAGUUAAGUGAACUGACCAUCGCGUCCAGC-------GAUUCUCGUUGGCGUUGCGGUGUUUUGUUUAUUUUGAGGCUAUAAUUUAGGCGGGUAAUUGCACGCUGGCGGCCAGAAU (((((.(((((((((((.((.(((((((.(((((-------(.....))))))...))))))).)).))))))))...............((((.((....)).)))))))..))))).. ( -38.10) >consensus UCUGGAGUUAAGUGAACUGACCAUC_____C______________AUCGUUGGCGUUGCGGUGUUUUGUUUAUUUUGAGGCUAUAAUUUAAGCGGGUAAUUGCACGCUGGCGGCCAGAAU (((((((((.....))))............................((((..((((.(((((..((((((((..(((......)))..))))))))..)))))))))..))))))))).. (-32.06 = -31.66 + -0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:22 2006