
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,656,670 – 9,656,867 |
| Length | 197 |
| Max. P | 0.956910 |

| Location | 9,656,670 – 9,656,763 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 83.38 |
| Mean single sequence MFE | -20.67 |
| Consensus MFE | -13.68 |
| Energy contribution | -14.65 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
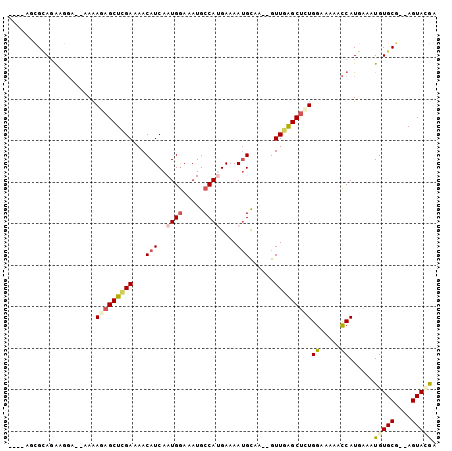
>3R_DroMel_CAF1 9656670 93 + 27905053 ----AGCGCAGAAGGA--AAAAGAGCUCGAAAACAUCAAUGGAAAUGCCACGAAAAUGCUA--GUUGAGCUUUGGAAAAACCAUGAAAUGUGCG--AGUACGA ----.(((((......--...(((((((((...(((...(((.....))).....)))...--.)))))))))((.....))......))))).--....... ( -20.20) >DroSec_CAF1 49248 97 + 1 CACAGUCGCAGAAGGA--AAAAGAGCUCGAAAACAUCAAUGGAAAUGCCAUGAAAAUGCUA--GUUGAGCUCUGGAAAAACCAUGAAAUGUGCG--AGUAAGA ...(.(((((.(....--...(((((((((...(((..((((.....))))....)))...--.)))))))))((.....))......).))))--).).... ( -25.60) >DroSim_CAF1 51676 90 + 1 -------GGAGAAGGA--AAAAGAGCUCGAAAACAUCAAUGGAAAUGCCAUGAAAAUGCUA--GUUGAGCUCUGGAAAAACCAUGAAAUGUGCG--AGUAAGA -------.........--......(((((...((((..((((.....(((.((....(((.--....)))))))).....))))...)))).))--))).... ( -19.10) >DroEre_CAF1 50306 93 + 1 ----AGUGCAGAAAGA--AAAAGAGCUGGAAAACAUCAAUGGAAAUGCCAUGAAAAUGCAG--GUUGAGCUGUGGAAAACCCAUGAAAUGUGCG--AGUACGA ----.(..((......--.....((((.((...(((..((((.....))))....)))...--.)).))))((((.....))))....))..).--....... ( -17.70) >DroWil_CAF1 61534 98 + 1 ----A-UGCAUGUGUAUGCAGACGGCUUGAAAACAUCAAUGAAAAUGCCAAGAAAAUGUAGUAGUUGGGCACUGGAAAAAUCAUGAAAUGUGCGCAAGUAUAU ----.-(((((....)))))...(((.(((.....)))((....)))))...........(((.(((.((((.................)))).))).))).. ( -18.73) >DroYak_CAF1 51455 93 + 1 ----AGCGUAGAAGGA--AAAAGAGCUCGAAAACAUCAAUGGAAAUGCCAUGAAAAGGCAG--GUUGAGCUCUGGAAAAUCCAUGAAAUGUGCG--AGUACGA ----..((((.(....--...(((((((((...(.((....))..((((.......)))))--.)))))))))((.....))......).))))--....... ( -22.70) >consensus ____AGCGCAGAAGGA__AAAAGAGCUCGAAAACAUCAAUGGAAAUGCCAUGAAAAUGCAA__GUUGAGCUCUGGAAAAACCAUGAAAUGUGCG__AGUACGA .....................(((((((((...(((..((((.....))))....)))......)))))))))((.....))......(((((....))))). (-13.68 = -14.65 + 0.97)


| Location | 9,656,696 – 9,656,799 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.05 |
| Mean single sequence MFE | -26.31 |
| Consensus MFE | -13.65 |
| Energy contribution | -14.07 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
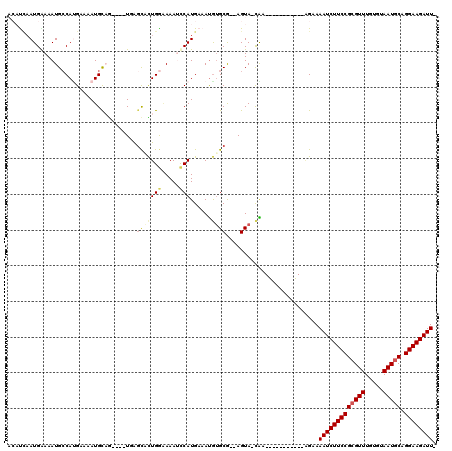
>3R_DroMel_CAF1 9656696 103 + 27905053 ACAUCAAUGGAAAUGCCACGAAAAUGCUA--GUUGAGCUUUGGAAAAACCAUGAAAUGUGCG--AGUA-CGA-----------GGAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUU- ((((..((((.....(((.......(((.--....)))..))).....))))...))))...--....-...-----------....(((((((((((((.....))))).))))))))- ( -22.90) >DroVir_CAF1 59761 110 + 1 --ACACAUGGAAAUGGCAUGAAAAUGCAG----UCAGCACUGGAAAAUCCAUGAAAUGUGCG--AGUA-CAAGUAUUAAAGCAAGAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUU- --...((((((....((((....))))..----(((....)))....)))))).....(((.--((((-....))))...)))....(((((((((((((.....))))).))))))))- ( -31.70) >DroGri_CAF1 63938 114 + 1 ACCUUAAUGAAAAUGCCAUGAAAAUGCUG----UUAGUACUGGCAAAUCCAUGAAAUGUGCG--AGUAGCAAGCAUUAAAGCAAGAAAAUCUUCCGCGUUUGUGUAAUUCAGGAAGAUUU ...((((((....((((((....)))...----....((((.(((.((.......)).))).--)))))))..)))))).......(((((((((..(((.....)))...))))))))) ( -22.90) >DroWil_CAF1 61561 107 + 1 ACAUCAAUGAAAAUGCCAAGAAAAUGUAGUAGUUGGGCACUGGAAAAAUCAUGAAAUGUGCGCAAGUA-UAU-----------GGCAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUU- .............(((((..........(((.(((.((((.................)))).))).))-).)-----------))))(((((((((((((.....))))).))))))))- ( -28.73) >DroMoj_CAF1 61469 110 + 1 --UCCAAUGAAAAUGCCAUGAAAAUGCAG----UCAACUUUGGGAAAUCCAUGAAAUGUGCG--AGUA-CAAGUAUUAAAGCAAGCAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUU- --((((((((...(((.((....))))).----)))...)))))..............(((.--((((-....))))...)))....(((((((((((((.....))))).))))))))- ( -26.50) >DroAna_CAF1 50949 103 + 1 ACAUCAAUGGAAAUGCCAUGAAAAUGCAG--GUUGAGCUCUGAAAAAUCCAUGAAACAAGCA--AGUA-CGG-----------UGAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUU- .((((.(((((.....(((....)))(((--(......)))).....)))))...((.....--.)).-.))-----------))..(((((((((((((.....))))).))))))))- ( -25.10) >consensus ACAUCAAUGAAAAUGCCAUGAAAAUGCAG____UGAGCACUGGAAAAUCCAUGAAAUGUGCG__AGUA_CAA___________AGAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUU_ ....................................(((((((.....)))......))))..........................(((((((((((((.....))))).)))))))). (-13.65 = -14.07 + 0.42)


| Location | 9,656,734 – 9,656,834 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.08 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -20.97 |
| Energy contribution | -21.17 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
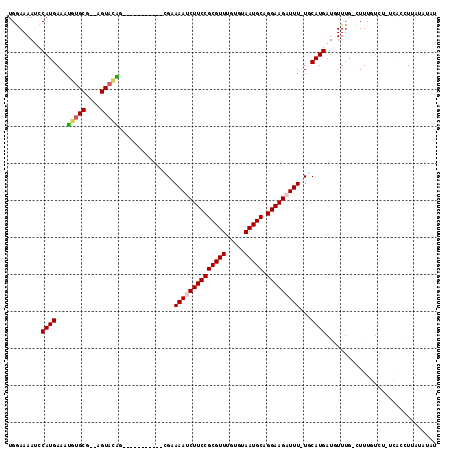
>3R_DroMel_CAF1 9656734 100 + 27905053 UGGAAAAACCAUGAAAUGUGCG--AGUACGA-----------GGAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUU-UCCAUGAUGUUUG-UUUUGUAU-UCGCUUUACAUUU (((.....)))..(((((((((--(((((((-----------(((((((((((((((((.....))))).)))))))))-)))..(((....)-))))))))-))))...)))))) ( -37.70) >DroVir_CAF1 59795 112 + 1 UGGAAAAUCCAUGAAAUGUGCG--AGUACAAGUAUUAAAGCAAGAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUU-UGCAUGAUGUUUG-CUUUUAUUUAGACAUUAUAUAU (((.....)))...(((((..(--((((.(((((......((.((((((((((((((((.....))))).)))))))))-).).)).....))-))).)))))..)))))...... ( -29.50) >DroPse_CAF1 49743 101 + 1 UGGAAAAUCCAUGAAAUGUGCG--AGUACAG-----------CGAAAACCUUCCGCGUUUGUGUAAUGCAGGAAGAUUU-UGCAUGAUGUUUGCCUUUGUCU-UCACCUCAUUCAC (((.....))).((...(.(((--(((((((-----------(((((..((((((((((.....))))).))))).)))-))).)).))))))))....)).-............. ( -23.20) >DroWil_CAF1 61601 103 + 1 UGGAAAAAUCAUGAAAUGUGCGCAAGUAUAU-----------GGCAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUUUGCAUGAUAUUUG-CUUUUUGU-UUUUUUGUUCAGU .(.(((.((((((..((((((....))))))-----------...((((((((((((((.....))))).)))))))))...)))))).))).-).......-............. ( -29.70) >DroAna_CAF1 50987 101 + 1 UGAAAAAUCCAUGAAACAAGCA--AGUACGG-----------UGAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUU-UGCAUGAUUUUCGCUUUUGUGU-UCGCCUUAAAUUU ......................--.....((-----------(((((((((((((((((.....))))).)))))))))-.(((..(.........)..)))-)))))........ ( -23.40) >DroPer_CAF1 49341 101 + 1 UGGAAAAUCCAUGAAAUGUGCG--AGUACAG-----------CGAAAACCUUCCGCGUUUGUGUAAUGCAGGAAGAUUU-UGCAUGAUGUUUGCCUUUGUCU-UCACCUCAUUCAC (((.....))).((...(.(((--(((((((-----------(((((..((((((((((.....))))).))))).)))-))).)).))))))))....)).-............. ( -23.20) >consensus UGGAAAAUCCAUGAAAUGUGCG__AGUACAG___________CGAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUU_UGCAUGAUGUUUG_CUUUGUCU_UCACCUUAUAUAU .........((((...(((((....)))))...............((((((((((((((.....))))).)))))))))...)))).............................. (-20.97 = -21.17 + 0.20)


| Location | 9,656,734 – 9,656,834 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.08 |
| Mean single sequence MFE | -22.83 |
| Consensus MFE | -12.54 |
| Energy contribution | -12.35 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9656734 100 - 27905053 AAAUGUAAAGCGA-AUACAAAA-CAAACAUCAUGGA-AAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUCC-----------UCGUACU--CGCACAUUUCAUGGUUUUUCCA ((((((...((((-.(((....-..........(((-(((((((((.((...........))))))))))))))-----------..))).)--)))))))))..(((.....))) ( -26.39) >DroVir_CAF1 59795 112 - 1 AUAUAUAAUGUCUAAAUAAAAG-CAAACAUCAUGCA-AAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUCUUGCUUUAAUACUUGUACU--CGCACAUUUCAUGGAUUUUCCA .........(((((....((((-(((.......(.(-(((((((((.((...........))))))))))))))))))))......(((...--.))).......)))))...... ( -18.31) >DroPse_CAF1 49743 101 - 1 GUGAAUGAGGUGA-AGACAAAGGCAAACAUCAUGCA-AAAUCUUCCUGCAUUACACAAACGCGGAAGGUUUUCG-----------CUGUACU--CGCACAUUUCAUGGAUUUUCCA ((((.((.(((((-((((....(((.......))).-....(((((.((...........))))))))))))))-----------)).)).)--)))........(((.....))) ( -26.40) >DroWil_CAF1 61601 103 - 1 ACUGAACAAAAAA-ACAAAAAG-CAAAUAUCAUGCAAAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUGCC-----------AUAUACUUGCGCACAUUUCAUGAUUUUUCCA .............-.......(-.(((.((((((...(((((((((.((...........)))))))))))(((-----------(......)).))......)))))).))).). ( -19.60) >DroAna_CAF1 50987 101 - 1 AAAUUUAAGGCGA-ACACAAAAGCGAAAAUCAUGCA-AAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUCA-----------CCGUACU--UGCUUGUUUCAUGGAUUUUUCA ...........((-(.(((..(((((..((..((.(-(((((((((.((...........))))))))))))))-----------..))..)--))))))))))............ ( -19.90) >DroPer_CAF1 49341 101 - 1 GUGAAUGAGGUGA-AGACAAAGGCAAACAUCAUGCA-AAAUCUUCCUGCAUUACACAAACGCGGAAGGUUUUCG-----------CUGUACU--CGCACAUUUCAUGGAUUUUCCA ((((.((.(((((-((((....(((.......))).-....(((((.((...........))))))))))))))-----------)).)).)--)))........(((.....))) ( -26.40) >consensus AUAUAUAAGGUGA_ACACAAAG_CAAACAUCAUGCA_AAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUCC___________CUGUACU__CGCACAUUUCAUGGAUUUUCCA .............................(((((...(((((((((.((...........)))))))))))................((......))......)))))........ (-12.54 = -12.35 + -0.19)



| Location | 9,656,763 – 9,656,867 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.53 |
| Mean single sequence MFE | -27.22 |
| Consensus MFE | -21.55 |
| Energy contribution | -21.80 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
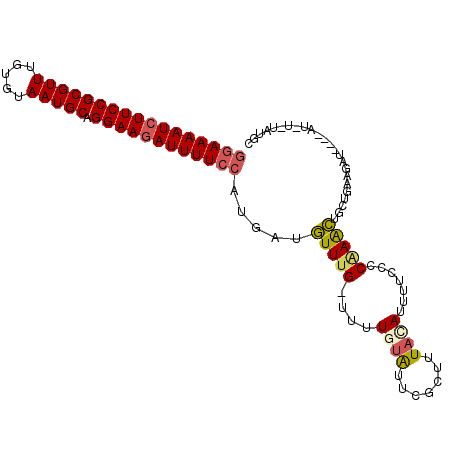
>3R_DroMel_CAF1 9656763 104 + 27905053 GGAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUUCCAUGAUGUUUG-UUUUGUAUUCGCUUUACAUUUUCCCCGAGCUGCUGAAAGUACAUAUGUAUAUGC (((((((((((((((((.....))))).))))))))))))...((((...-(((.(((((((..............)))).))).)))...)))).......... ( -29.04) >DroSec_CAF1 49345 100 + 1 GGAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUUCCACGUUGUUUG-UUUUCUAUACGCUUUACAUUUUCCCCAAGCGGCUGAAAAU----AUAUUUAUGC (((((((((((((((((.....))))).)))))))))))).(((.((.((-(((((....(((((............)))))...))))))----).))..))). ( -32.90) >DroSim_CAF1 51766 96 + 1 GGAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUUCCAUGAUGUUUG-UUUUGUAUUCGCUUUACAUUUUCCCCAAGCGGCUGAAGAU----AU----AUGC (((((((((((((((((.....))))).))))))))))))........((-((((.((..(((((............)))))..)))))))----).----.... ( -29.30) >DroEre_CAF1 50399 99 + 1 GGAAAAU-UUCCGCGUUUGUGUAAUGCAGGAAGAUUUUCCAUGAUGUUUG-UUUUGUAUUCGCUUUAUAUACUCCCCGAACUGCUGAAGAC----AUUUGUAUGC (((((((-(((((((((.....))))).)).)))))))))..(((((((.-....(((((((..............)))).)))...))))----)))....... ( -22.44) >DroYak_CAF1 51548 100 + 1 GGAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUUCCAUGAUGUUUG-UUUUGUAUUCGCUUUUCAUUUUCCCCGAACUGCUGAAGAC----AUUUGUAUGC (((((((((((((((((.....))))).))))))))))))..(((((((.-....(((((((..............)))).)))...))))----)))....... ( -29.74) >DroAna_CAF1 51016 81 + 1 UGAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUUGCAUGAUUUUCGCUUUUGUGUUCGCCUUAAAUUUGCCACAAAA------------------------ (((((((((((((((((.....))))).)))))))))).))...........((((((...((.........)))))))).------------------------ ( -19.90) >consensus GGAAAAUCUUCCGCGUUUGUGUAAUGCAGGAAGAUUUUCCAUGAUGUUUG_UUUUGUAUUCGCUUUACAUUUUCCCCAAACUGCUGAAGAU____AU_U_UAUGC (((((((((((((((((.....))))).)))))))))))).....(((((....((((.......)))).......)))))........................ (-21.55 = -21.80 + 0.25)

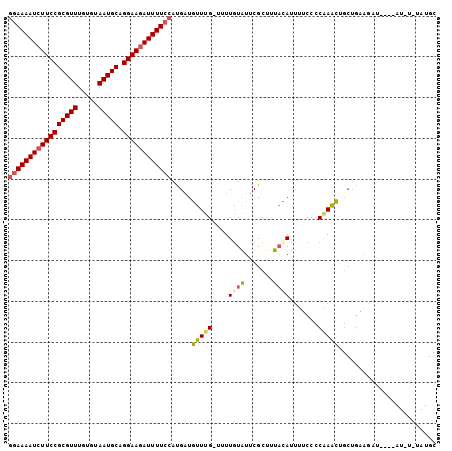
| Location | 9,656,763 – 9,656,867 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.53 |
| Mean single sequence MFE | -21.30 |
| Consensus MFE | -15.22 |
| Energy contribution | -15.72 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9656763 104 - 27905053 GCAUAUACAUAUGUACUUUCAGCAGCUCGGGGAAAAUGUAAAGCGAAUACAAAA-CAAACAUCAUGGAAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUCC ((...(((((......((((..(.....)..)))))))))..))..........-..........((((((((((((.((...........)))))))))))))) ( -21.00) >DroSec_CAF1 49345 100 - 1 GCAUAAAUAU----AUUUUCAGCCGCUUGGGGAAAAUGUAAAGCGUAUAGAAAA-CAAACAACGUGGAAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUCC .......(((----((((((..((....)).)))))))))..((((........-......))))((((((((((((.((...........)))))))))))))) ( -27.24) >DroSim_CAF1 51766 96 - 1 GCAU----AU----AUCUUCAGCCGCUUGGGGAAAAUGUAAAGCGAAUACAAAA-CAAACAUCAUGGAAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUCC ((.(----((----((.(((..((....)).))).)))))..))..........-..........((((((((((((.((...........)))))))))))))) ( -23.80) >DroEre_CAF1 50399 99 - 1 GCAUACAAAU----GUCUUCAGCAGUUCGGGGAGUAUAUAAAGCGAAUACAAAA-CAAACAUCAUGGAAAAUCUUCCUGCAUUACACAAACGCGGAA-AUUUUCC ((........----.(((((........))))).........))..........-..........(((((((.((((.((...........))))))-))))))) ( -13.77) >DroYak_CAF1 51548 100 - 1 GCAUACAAAU----GUCUUCAGCAGUUCGGGGAAAAUGAAAAGCGAAUACAAAA-CAAACAUCAUGGAAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUCC .........(----((.(((.((..((((.......))))..))))).)))...-..........((((((((((((.((...........)))))))))))))) ( -24.90) >DroAna_CAF1 51016 81 - 1 ------------------------UUUUGUGGCAAAUUUAAGGCGAACACAAAAGCGAAAAUCAUGCAAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUCA ------------------------(((((((................)))))))(((.......)))((((((((((.((...........)))))))))))).. ( -17.09) >consensus GCAUA_A_AU____AUCUUCAGCAGCUCGGGGAAAAUGUAAAGCGAAUACAAAA_CAAACAUCAUGGAAAAUCUUCCUGCAUUACACAAACGCGGAAGAUUUUCC .................................................................((((((((((((.((...........)))))))))))))) (-15.22 = -15.72 + 0.50)


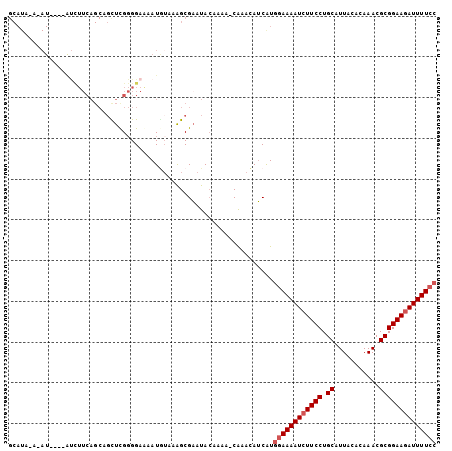
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:20 2006