| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,644,742 – 9,644,845 |
| Length | 103 |
| Max. P | 0.863807 |

| Location | 9,644,742 – 9,644,845 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 76.55 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -14.34 |
| Energy contribution | -14.20 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
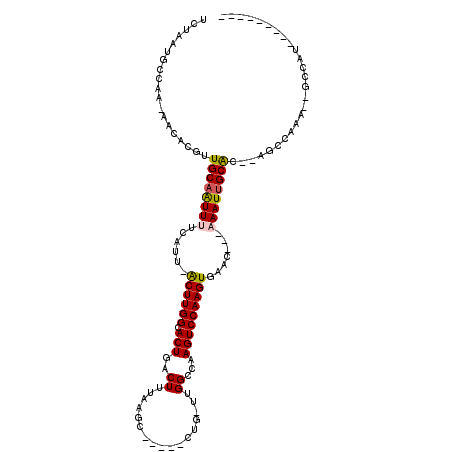
>3R_DroMel_CAF1 9644742 103 + 27905053 UCUAAUGCCAA-AACACGUUGCAAUUUUCAUU-GCUUGGCACUGACUUUAGGC-----UUG-UUGGCCAAGUCCAAGUUAAC---CAAUUGCGC--AGCCAGU--GCCAUU-UGGCGAG .....((((((-(.......(((((....)))-)).((((((((.((...(((-----(..-..))))..((.(((......---...))).))--)).))))--))))))-))))).. ( -33.30) >DroVir_CAF1 43600 98 + 1 UCUAAUGCCAA-AACACGUUGCAAUUUUCAUU-ACUUGGCACUGACUUUAAUGC----CUC-UUGGCCAAGUCCAAGUGAAC---AAAUUGCAGCCAGCCAAA--GCCGU--------- ......((...-.....((((((((((...((-((((((.(((.........((----(..-..)))..)))))))))))..---))))))))))........--))...--------- ( -25.69) >DroGri_CAF1 50516 109 + 1 UCUAAUGCCAAAAACACGUUGCAGUUUUCAUUCACUUGGCACUGACUUUAAGCCGCUGCUG-UUGGCCAAGUCCAAGUGAACAAAAAAUUGCACUUAGCCAAAAAGACGU--------- ...............(((((((((((((..(((((((((.(((..((...(((....))).-..))...))))))))))))...)))))))))(((.......)))))))--------- ( -28.20) >DroWil_CAF1 40359 105 + 1 UCUAAUGCCAA-AACACGUUGCCAUUUUCAUU-ACUUGGCACUGACUUUAAGC-----UGGCUUGGCCAAGUCCAAGUUAAC---AAAUUGCGC--AGCCAUA--GCCACUAAAUACAU ...........-....((.(((((........-...))))).))......((.-----(((((((((...((.(((......---...))).))--.)))).)--))))))........ ( -20.90) >DroMoj_CAF1 46519 103 + 1 UCUAAUGCCAA-AGCACGUUGCAAUUUUCAUU-GCUUGGCACUGACUUUAAAGC--GACUG-UUGGCCAAGUCCAAGUGAACAACAAAUUGCAGCUAGAUAAA--GUGCU--------- ...........-((((((((((((((((((((-(((((((...(((........--....)-)).)))))))...))))).....))))))))))........--)))))--------- ( -27.50) >DroAna_CAF1 36976 97 + 1 UCUAAUGCCAA-AACACGUUGCAAUUUUCAUG-GCUUGGCACUGACUUUAGGC-----CUG-UUGGCCAAGUCCAAGUUAAC---CAAUUGCGC--AGCC--------AUU-CAGCCCG ...........-.....((.((((((....((-((((((.(((.......(((-----(..-..)))).)))))))))))..---.))))))))--....--------...-....... ( -23.20) >consensus UCUAAUGCCAA_AACACGUUGCAAUUUUCAUU_ACUUGGCACUGACUUUAAGC_____CUG_UUGGCCAAGUCCAAGUGAAC___AAAUUGCAC__AGCCAAA__GCCAU_________ ...................((((((((......((((((.(((..((.................))...))))))))).......)))))))).......................... (-14.34 = -14.20 + -0.14)

| Location | 9,644,742 – 9,644,845 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 76.55 |
| Mean single sequence MFE | -31.45 |
| Consensus MFE | -14.46 |
| Energy contribution | -14.99 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
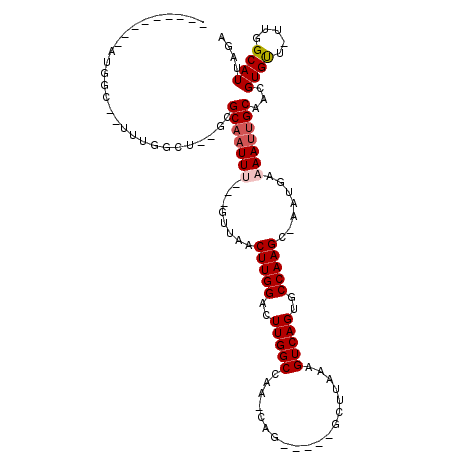
>3R_DroMel_CAF1 9644742 103 - 27905053 CUCGCCA-AAUGGC--ACUGGCU--GCGCAAUUG---GUUAACUUGGACUUGGCCAA-CAA-----GCCUAAAGUCAGUGCCAAGC-AAUGAAAAUUGCAACGUGUU-UUGGCAUUAGA ...((((-((((((--(((((((--..((..(((---(((((.......))))))))-...-----))....))))))))))).((-(((....))))).......)-)))))...... ( -42.30) >DroVir_CAF1 43600 98 - 1 ---------ACGGC--UUUGGCUGGCUGCAAUUU---GUUCACUUGGACUUGGCCAA-GAG----GCAUUAAAGUCAGUGCCAAGU-AAUGAAAAUUGCAACGUGUU-UUGGCAUUAGA ---------.((((--(......)))))......---((((....))))...(((((-(((----(((((......))))))..((-(((....)))))......))-)))))...... ( -29.20) >DroGri_CAF1 50516 109 - 1 ---------ACGUCUUUUUGGCUAAGUGCAAUUUUUUGUUCACUUGGACUUGGCCAA-CAGCAGCGGCUUAAAGUCAGUGCCAAGUGAAUGAAAACUGCAACGUGUUUUUGGCAUUAGA ---------..(((.....(((....((((..((((..(((((((((..(((((...-.(((....)))....)))))..)))))))))..)))).))))....)))...)))...... ( -33.90) >DroWil_CAF1 40359 105 - 1 AUGUAUUUAGUGGC--UAUGGCU--GCGCAAUUU---GUUAACUUGGACUUGGCCAAGCCA-----GCUUAAAGUCAGUGCCAAGU-AAUGAAAAUGGCAACGUGUU-UUGGCAUUAGA ...........(((--(..((((--(.((.....---(....)((((......))))))))-----)))...))))(((((((((.-.......(((....)))..)-))))))))... ( -26.70) >DroMoj_CAF1 46519 103 - 1 ---------AGCAC--UUUAUCUAGCUGCAAUUUGUUGUUCACUUGGACUUGGCCAA-CAGUC--GCUUUAAAGUCAGUGCCAAGC-AAUGAAAAUUGCAACGUGCU-UUGGCAUUAGA ---------.(.((--((((...(((.((....(((((..((........))..)))-)))).--))).)))))))((((((((((-(((....)))))........-))))))))... ( -27.40) >DroAna_CAF1 36976 97 - 1 CGGGCUG-AAU--------GGCU--GCGCAAUUG---GUUAACUUGGACUUGGCCAA-CAG-----GCCUAAAGUCAGUGCCAAGC-CAUGAAAAUUGCAACGUGUU-UUGGCAUUAGA ..((((.-...--------((((--......(((---(((((.......))))))))-..)-----)))...))))(((((((((.-((((..........)))).)-))))))))... ( -29.20) >consensus _________AUGGC__UUUGGCU__GCGCAAUUU___GUUAACUUGGACUUGGCCAA_CAG_____GCUUAAAGUCAGUGCCAAGC_AAUGAAAAUUGCAACGUGUU_UUGGCAUUAGA ...........................(((((((........(((((..(((((...................)))))..))))).......)))))))...((((.....)))).... (-14.46 = -14.99 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:13 2006