| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
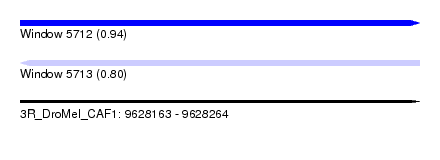
| Location | 9,628,163 – 9,628,264 |
| Length | 101 |
| Max. P | 0.944802 |

| Location | 9,628,163 – 9,628,264 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 86.85 |
| Mean single sequence MFE | -34.95 |
| Consensus MFE | -28.63 |
| Energy contribution | -29.55 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
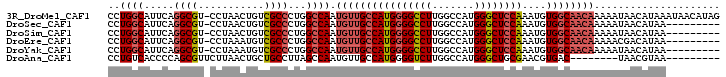
>3R_DroMel_CAF1 9628163 101 + 27905053 CCUGGCAUUCAGGCGU-CCUAACUGUCGCCCUGGCCAAUGUUGCCAUGGGGCCUUGGCCAUGGGCUCCAAAUGUGGCAACAAAAAUAACAUAAAUAACAUAG ..((((.....((((.-(......).))))...)))).((((((((((((((((.......))))))))....))))))))..................... ( -37.00) >DroSec_CAF1 21149 92 + 1 CCUGGCAUUCAGGCGU-CCUAACUGUCGCCCUGGCCAAUGUUGCCAUGGGGCCUUGGCCAUGGGCUCCAAAUGUGGCAACAAAAAUAACAUAA--------- ..((((.....((((.-(......).))))...)))).((((((((((((((((.......))))))))....))))))))............--------- ( -37.00) >DroSim_CAF1 22410 92 + 1 CCUGGCAUUCAGGCGU-CCUAACUGUCGCCCUGGCCAAUGUUGCCAUGGGGCCUUGGCCAUGGGCUCCAAAUGUGGCAACAAAAAUAACAUAA--------- ..((((.....((((.-(......).))))...)))).((((((((((((((((.......))))))))....))))))))............--------- ( -37.00) >DroEre_CAF1 21648 92 + 1 CCUGGCAUUCAGGCGU-CCUAAAUGUCGCCCUGGCCAAUGUUGCCAUGGGGCCUUGGCCAUGGGCUCCAAAUGUGGCAACAAAAACGACAUAA--------- ..((((.....((((.-(......).))))...)))).((((((((((((((((.......))))))))....))))))))............--------- ( -37.00) >DroYak_CAF1 21864 92 + 1 CCUGGCAUUCAGGCGU-CCUAAAUGUCGCCCUGGCCAAUGUUGCCAUGGGGCCUUGGCCAUGGGCUCCAAAUGUGGCAACAAAAAUAACAUAA--------- ..((((.....((((.-(......).))))...)))).((((((((((((((((.......))))))))....))))))))............--------- ( -37.00) >DroAna_CAF1 21089 85 + 1 CCUGUCACCCCAGCGUUCUUAACUGCUGCCUUAGCCAAUGUUGCCAUGGGGUCUUGGCCAUGGGCUGCGAACGUGAC--------UAACGUAA--------- ...(((((..(((((........))))).(.(((((.(((..((((........))))))).))))).)...)))))--------........--------- ( -24.70) >consensus CCUGGCAUUCAGGCGU_CCUAACUGUCGCCCUGGCCAAUGUUGCCAUGGGGCCUUGGCCAUGGGCUCCAAAUGUGGCAACAAAAAUAACAUAA_________ ..((((.....((((...........))))...)))).((((((((((((((((.......))))))))....))))))))..................... (-28.63 = -29.55 + 0.92)

| Location | 9,628,163 – 9,628,264 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 86.85 |
| Mean single sequence MFE | -34.24 |
| Consensus MFE | -28.02 |
| Energy contribution | -28.30 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.803867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9628163 101 - 27905053 CUAUGUUAUUUAUGUUAUUUUUGUUGCCACAUUUGGAGCCCAUGGCCAAGGCCCCAUGGCAACAUUGGCCAGGGCGACAGUUAGG-ACGCCUGAAUGCCAGG .............(((....(((((.(((....))).((((.(((((((.(((....)))....))))))))))))))))....)-)).((((.....)))) ( -35.70) >DroSec_CAF1 21149 92 - 1 ---------UUAUGUUAUUUUUGUUGCCACAUUUGGAGCCCAUGGCCAAGGCCCCAUGGCAACAUUGGCCAGGGCGACAGUUAGG-ACGCCUGAAUGCCAGG ---------....(((....(((((.(((....))).((((.(((((((.(((....)))....))))))))))))))))....)-)).((((.....)))) ( -35.70) >DroSim_CAF1 22410 92 - 1 ---------UUAUGUUAUUUUUGUUGCCACAUUUGGAGCCCAUGGCCAAGGCCCCAUGGCAACAUUGGCCAGGGCGACAGUUAGG-ACGCCUGAAUGCCAGG ---------....(((....(((((.(((....))).((((.(((((((.(((....)))....))))))))))))))))....)-)).((((.....)))) ( -35.70) >DroEre_CAF1 21648 92 - 1 ---------UUAUGUCGUUUUUGUUGCCACAUUUGGAGCCCAUGGCCAAGGCCCCAUGGCAACAUUGGCCAGGGCGACAUUUAGG-ACGCCUGAAUGCCAGG ---------..((((.((.......)).))))((((.((((.(((((((.(((....)))....)))))))))))..((((((((-...)))))))))))). ( -37.10) >DroYak_CAF1 21864 92 - 1 ---------UUAUGUUAUUUUUGUUGCCACAUUUGGAGCCCAUGGCCAAGGCCCCAUGGCAACAUUGGCCAGGGCGACAUUUAGG-ACGCCUGAAUGCCAGG ---------.......................((((.((((.(((((((.(((....)))....)))))))))))..((((((((-...)))))))))))). ( -35.60) >DroAna_CAF1 21089 85 - 1 ---------UUACGUUA--------GUCACGUUCGCAGCCCAUGGCCAAGACCCCAUGGCAACAUUGGCUAAGGCAGCAGUUAAGAACGCUGGGGUGACAGG ---------........--------(((((.(..((.(((..((((((.......(((....))))))))).)))....((.....))))..).)))))... ( -25.61) >consensus _________UUAUGUUAUUUUUGUUGCCACAUUUGGAGCCCAUGGCCAAGGCCCCAUGGCAACAUUGGCCAGGGCGACAGUUAGG_ACGCCUGAAUGCCAGG ................................((((.((((.(((((((.(((....)))....)))))))))))..((.(((((....))))).)))))). (-28.02 = -28.30 + 0.28)

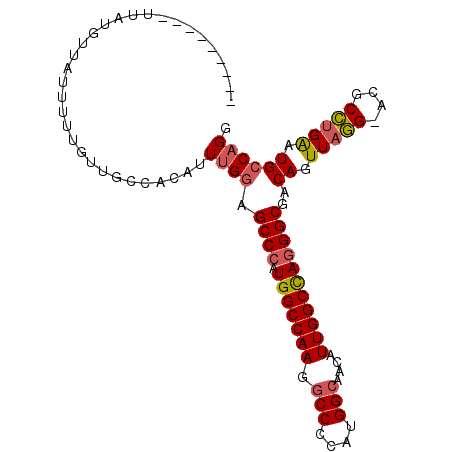
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:07 2006