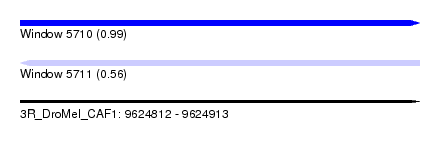
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,624,812 – 9,624,913 |
| Length | 101 |
| Max. P | 0.987356 |

| Location | 9,624,812 – 9,624,913 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 88.20 |
| Mean single sequence MFE | -38.88 |
| Consensus MFE | -31.10 |
| Energy contribution | -32.83 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.04 |
| Mean z-score | -4.30 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9624812 101 + 27905053 AUCCAGGCAGCUGCAAGCCA-CAAACUA-ACAUGCAAAGGAUCUCGGCCGGAUCUGGUAUUGAAUCCAAUCUCAAUCCGGCGGUGUCCAACAACAGCUGCCAA .....((((((((...((..-.......-....))...((((..(.(((((((.((..((((....))))..))))))))).).)))).....)))))))).. ( -39.32) >DroSec_CAF1 17789 101 + 1 AUCCAGGCAGCUGCAAGCCG-CAAACUA-ACAUGCAAAGGAUUUCGGCCGGAUCUGGUAUUGAAUCCAAUCUCGAUCCGGCGGUGUCCAACAACAGCUGCCAA .....((((((((......(-((.....-...)))...((((..(.((((((((.((.((((....)))))).)))))))).).)))).....)))))))).. ( -45.20) >DroSim_CAF1 18852 101 + 1 GCCCAGGCAGCUGCAAGCCA-CAAACUA-ACAUGCAAAGGAUCUCGGCCGGAUCUGGUAUUGAAUCCAAUCUCGAUCCGGUGGUGUCCAACAACAGCUGCCAA .....((((((((...((..-.......-....))...((((..(.((((((((.((.((((....)))))).)))))))).).)))).....)))))))).. ( -40.92) >DroEre_CAF1 18263 101 + 1 AUCUCGGCAGCUGCAAGCCA-CAAACUA-ACAUGCAAAGGAUCUCGGCCGGAUCUGGUAUUGAAUCCAAUCUCGUUCCGGCGGUGUCCAACAACAGCUGCCAA .....((((((((...((..-.......-....))...((((..(.((((((.(.((.((((....)))))).).)))))).).)))).....)))))))).. ( -37.82) >DroYak_CAF1 18336 101 + 1 AUACCGGCAGCUGCAAGCCA-CAAACUA-AAAUGCAAAGGAUCUCGGCCGGAUCUGGUAUUGAAUCCAAUCUCGUUCCGGCGGUGUCCAACAACAGCUGCCAA .....((((((((...((..-.......-....))...((((..(.((((((.(.((.((((....)))))).).)))))).).)))).....)))))))).. ( -37.82) >DroAna_CAF1 17731 92 + 1 AUCCAGGCAGCUGCAAACUAUCAGACCAUAUAUGCAAAGGAUCUGGGCCGGA-----------AUUGAAUCUCGUUCCGGAGGUGUCCAACGACAGCUGCCAG .....(((((((((......(((((((...........)).))))).(((((-----------((........)))))))...........).)))))))).. ( -32.20) >consensus AUCCAGGCAGCUGCAAGCCA_CAAACUA_ACAUGCAAAGGAUCUCGGCCGGAUCUGGUAUUGAAUCCAAUCUCGAUCCGGCGGUGUCCAACAACAGCUGCCAA .....((((((((...((...............))...((((..(.((((((.(.((.((((....)))))).).)))))).).)))).....)))))))).. (-31.10 = -32.83 + 1.72)


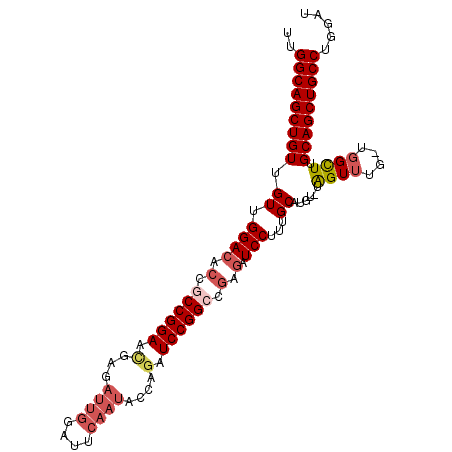
| Location | 9,624,812 – 9,624,913 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 88.20 |
| Mean single sequence MFE | -37.17 |
| Consensus MFE | -29.09 |
| Energy contribution | -30.37 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9624812 101 - 27905053 UUGGCAGCUGUUGUUGGACACCGCCGGAUUGAGAUUGGAUUCAAUACCAGAUCCGGCCGAGAUCCUUUGCAUGU-UAGUUUG-UGGCUUGCAGCUGCCUGGAU ..(((((((((.(((((((.(.((((((((...((((....))))....)))))))).).).)))...(((...-.....))-))))..)))))))))..... ( -39.00) >DroSec_CAF1 17789 101 - 1 UUGGCAGCUGUUGUUGGACACCGCCGGAUCGAGAUUGGAUUCAAUACCAGAUCCGGCCGAAAUCCUUUGCAUGU-UAGUUUG-CGGCUUGCAGCUGCCUGGAU ..(((((((((.((((((..(.((((((((...((((....))))....)))))))).)...)))...(((...-.....))-))))..)))))))))..... ( -42.10) >DroSim_CAF1 18852 101 - 1 UUGGCAGCUGUUGUUGGACACCACCGGAUCGAGAUUGGAUUCAAUACCAGAUCCGGCCGAGAUCCUUUGCAUGU-UAGUUUG-UGGCUUGCAGCUGCCUGGGC ..(((((((((.(((((((.(..(((((((...((((....))))....)))))))..).).)))...(((...-.....))-))))..)))))))))..... ( -36.50) >DroEre_CAF1 18263 101 - 1 UUGGCAGCUGUUGUUGGACACCGCCGGAACGAGAUUGGAUUCAAUACCAGAUCCGGCCGAGAUCCUUUGCAUGU-UAGUUUG-UGGCUUGCAGCUGCCGAGAU (((((((((((.(((((((.(.((((((.(...((((....))))....).)))))).).).)))...(((...-.....))-))))..)))))))))))... ( -38.00) >DroYak_CAF1 18336 101 - 1 UUGGCAGCUGUUGUUGGACACCGCCGGAACGAGAUUGGAUUCAAUACCAGAUCCGGCCGAGAUCCUUUGCAUUU-UAGUUUG-UGGCUUGCAGCUGCCGGUAU (((((((((((.(((((((.(.((((((.(...((((....))))....).)))))).).).)))...(((...-.....))-))))..)))))))))))... ( -37.00) >DroAna_CAF1 17731 92 - 1 CUGGCAGCUGUCGUUGGACACCUCCGGAACGAGAUUCAAU-----------UCCGGCCCAGAUCCUUUGCAUAUAUGGUCUGAUAGUUUGCAGCUGCCUGGAU ..(((((((((....((((....((((((..........)-----------)))))..((((.((...........))))))...)))))))))))))..... ( -30.40) >consensus UUGGCAGCUGUUGUUGGACACCGCCGGAACGAGAUUGGAUUCAAUACCAGAUCCGGCCGAGAUCCUUUGCAUGU_UAGUUUG_UGGCUUGCAGCUGCCUGGAU ..(((((((((.((.((((.(.((((((.(...((((....))))....).)))))).).).)))...))......((((....)))).)))))))))..... (-29.09 = -30.37 + 1.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:05 2006