
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,597,487 – 9,597,632 |
| Length | 145 |
| Max. P | 0.987301 |

| Location | 9,597,487 – 9,597,606 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.28 |
| Mean single sequence MFE | -28.25 |
| Consensus MFE | -25.71 |
| Energy contribution | -25.44 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9597487 119 + 27905053 CACAGACGACCCAAACGAAAAGAAAAGACGACCACGACGGCCAGAAAUAAGCAA-AUUAUUUGGCUAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAA .............................((((.....((((((((((......-))).)))))))(((((((((((...(((.............)))...)))))))))))))))... ( -30.22) >DroPse_CAF1 5117 119 + 1 CACAGACGACGAAUACGACGACU-UCGAACACGACAACGGCCACAAAUAAGCAAAAUUAUUUGGCUAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGAUGAUUGUUGGUUUAA ..................((...-(((....)))...))....(((((((......)))))))((((((((((((((...(((.............)))...)))))))))))))).... ( -28.22) >DroWil_CAF1 4790 96 + 1 CACAGACGACG----------------ACGA--------GCCAAAAAUAAGCAAAAUUAUUUAGCUGACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAA ...........----------------....--------....................((((((..((((((((((...(((.............)))...))))))))))..)))))) ( -26.52) >DroYak_CAF1 4956 114 + 1 CACAGACGACCAA-----AACGAAAAGACGACCACGACGGCCAGAAAUAAGCAA-AUUAUUUAGCUAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAA .............-----...........(.((.....)).)............-....((((((((((((((((((...(((.............)))...)))))))))))))))))) ( -27.82) >DroAna_CAF1 4844 97 + 1 CACAGAC----------------------GACCACGACGGCCAGAAAUAAGCAA-AUUAUUUAGCUAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAA .......----------------------(.((.....)).)............-....((((((((((((((((((...(((.............)))...)))))))))))))))))) ( -27.82) >DroPer_CAF1 5047 119 + 1 CACAGACGACGAAUACGACGACU-UCGACCACGACAACGGCCAGAAAUAAGCAAAAUUAUUUGGCUAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGAUGAUUGUUGGUUUAA ......((.((....)).))...-..((((........(((((..(((((......))))))))))(((((((((((...(((.............)))...)))))))))))))))... ( -28.92) >consensus CACAGACGACGAA_______A_____GACGACCACGACGGCCAGAAAUAAGCAA_AUUAUUUAGCUAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAA ...........................................................((((((((((((((((((...(((.............)))...)))))))))))))))))) (-25.71 = -25.44 + -0.28)

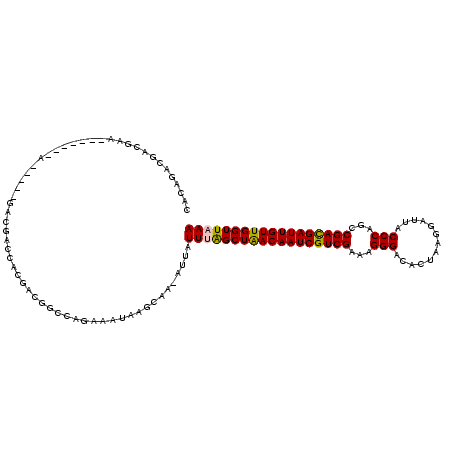

| Location | 9,597,527 – 9,597,632 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.68 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -29.00 |
| Energy contribution | -28.64 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.16 |
| Mean z-score | -4.09 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9597527 105 + 27905053 CCAGAAAUAAGCAA-AUUAUUUGGCUAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAAUUUUGCCAACGGC-----AGCGCGAA----AACUU ..........((..-...(((((((((((((((((((...(((.............)))...))))))))))))))))))).(((((...)))-----)).))...----..... ( -34.32) >DroVir_CAF1 4110 98 + 1 CCACAAAUAAGCAA-AUUAUUUAGCUGACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUCCGUUAAAGUUUGCGAGCGGCUAGCUA---------------- ..........((((-((..((((((.(((((((((((...(((.............)))...))))))))))).)))))))))))).(((.....))).---------------- ( -34.72) >DroPse_CAF1 5156 101 + 1 CCACAAAUAAGCAAAAUUAUUUGGCUAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGAUGAUUGUUGGUUUAAAUUUGCUAACGGC-----ACCGCGAA--------- .........((((((.(((...(((((((((((((((...(((.............)))...)))))))))))))))))).))))))..((..-----..))....--------- ( -29.02) >DroWil_CAF1 4806 92 + 1 CCAAAAAUAAGCAAAAUUAUUUAGCUGACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAAAAUUGCAAAA--------------AA--------- ..........((((.....((((((..((((((((((...(((.............)))...))))))))))..))))))..))))....--------------..--------- ( -29.92) >DroMoj_CAF1 3944 114 + 1 CCAGAAAUAAGCAA-AUUAUUUAGCUGACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAAAUUUGCGAAAGGCUAGCUAACGCGAACUACAAAUU ..((......((((-(((..(((((..((((((((((...(((.............)))...))))))))))..))))))))))))......)).((....))............ ( -36.52) >DroPer_CAF1 5086 101 + 1 CCAGAAAUAAGCAAAAUUAUUUGGCUAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGAUGAUUGUUGGUUUAAAUUUGCUAACGGC-----AGCGCGAA--------- .........((((((.(((...(((((((((((((((...(((.............)))...)))))))))))))))))).))))))......-----........--------- ( -28.42) >consensus CCAGAAAUAAGCAA_AUUAUUUAGCUAACAAUCGUCGAAAGGGACACUAAGGAUUACCCAGCCGACGAUUGUUGGUUAAAAUUUGCGAACGGC_____A_CGCGAA_________ ..........((((.....((((((((((((((((((...(((.............)))...))))))))))))))))))..))))............................. (-29.00 = -28.64 + -0.36)



| Location | 9,597,527 – 9,597,632 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.68 |
| Mean single sequence MFE | -26.04 |
| Consensus MFE | -21.10 |
| Energy contribution | -21.60 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9597527 105 - 27905053 AAGUU----UUCGCGCU-----GCCGUUGGCAAAAUUUAACCAACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUUAGCCAAAUAAU-UUGCUUAUUUCUGG .....----...(((..-----....(((((...........((((((((((((..(((.............)))..)))))))))))).))))).....-.))).......... ( -27.52) >DroVir_CAF1 4110 98 - 1 ----------------UAGCUAGCCGCUCGCAAACUUUAACGGACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUCAGCUAAAUAAU-UUGCUUAUUUGUGG ----------------.(((.....)))((((((........((((((((((((..(((.............)))..))))))))))))(((........-..)))..)))))). ( -31.02) >DroPse_CAF1 5156 101 - 1 ---------UUCGCGGU-----GCCGUUAGCAAAUUUAAACCAACAAUCAUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUUAGCCAAAUAAUUUUGCUUAUUUGUGG ---------..(((((.-----......((((((.(((..(.(((((((.((((..(((.............)))..)))).))))))).).....))).))))))...))))). ( -22.82) >DroWil_CAF1 4806 92 - 1 ---------UU--------------UUUUGCAAUUUUUAACCAACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUCAGCUAAAUAAUUUUGCUUAUUUUUGG ---------..--------------....((((..((((.(..(((((((((((..(((.............)))..)))))))))))..).)))).....)))).......... ( -24.42) >DroMoj_CAF1 3944 114 - 1 AAUUUGUAGUUCGCGUUAGCUAGCCUUUCGCAAAUUUUAACCAACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUCAGCUAAAUAAU-UUGCUUAUUUCUGG ........(((.((....)).))).....((((((((((.(..(((((((((((..(((.............)))..)))))))))))..).)))..)))-)))).......... ( -29.62) >DroPer_CAF1 5086 101 - 1 ---------UUCGCGCU-----GCCGUUAGCAAAUUUAAACCAACAAUCAUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUUAGCCAAAUAAUUUUGCUUAUUUCUGG ---------...(((((-----(....))))...........(((((((.((((..(((.............)))..)))).)))))))..............)).......... ( -20.82) >consensus _________UUCGCG_U_____GCCGUUAGCAAAUUUUAACCAACAAUCGUCGGCUGGGUAAUCCUUAGUGUCCCUUUCGACGAUUGUCAGCCAAAUAAU_UUGCUUAUUUCUGG .............................((((.........((((((((((((..(((.............)))..))))))))))))............)))).......... (-21.10 = -21.60 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:56 2006