| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,376,912 – 1,377,019 |
| Length | 107 |
| Max. P | 0.929830 |

| Location | 1,376,912 – 1,377,019 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.17 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -13.44 |
| Energy contribution | -13.56 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.96 |
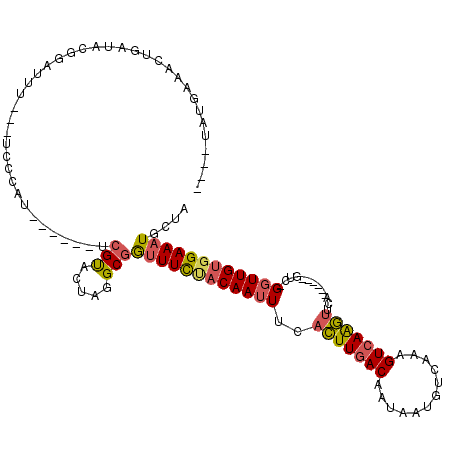
| Structure conservation index | 0.55 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
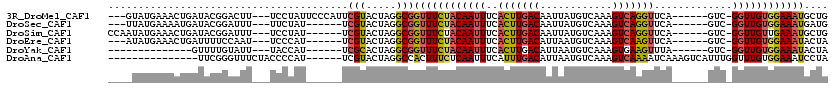
>3R_DroMel_CAF1 1376912 107 - 27905053 ---GUAUGAAACUGAUACGGACUU---UCCUAUUCCCAUUCGUACUAGGCGGUUUCUACAAUUUCACUUGACAAUUAUGUCAAAGUCAGGUUCA------GUC-GGUUGUGGAAAUGCUG ---(((((((...((((.((....---.))))))....)))))))..((((.(((((((((((..(((((((............)))))))...------...-))))))))))))))). ( -28.60) >DroSec_CAF1 3037 101 - 1 ---UUAUGAAAAUGAUACGGAUUU---UUCUAU------UCGUACUAGGCGGUUUCUACAAUUUCACUUGACAAUUAUGUCAAAGUCAGGUUCA------GUC-GGUUGUGGAAAUGAUG ---((((....))))(((((((..---....))------))))).......((((((((((((..(((((((............)))))))...------...-)))))))))))).... ( -23.40) >DroSim_CAF1 3046 104 - 1 CCAAUAUGAAACUGAUACGGAUUU---UCCUAU------UCGUACUAGGCGGUUUCUACAAUUUCACUUGACAAUUAUGUCAAAGUCAGGUUCA------GUC-GGUUGUUGAAAUGCUG ...........(((.(((((((..---....))------))))).)))(((.((((.((((((..(((((((............)))))))...------...-)))))).))))))).. ( -22.90) >DroEre_CAF1 3179 101 - 1 ---AUAUGAAACUGAUUUUCCAAU---UCCCAU------UCGUACUAGGCGGUUUCUACAAUUUCACUUGACAUUAAUGUCAAAGUCAAGUUCA------GUC-GGUUGUGGAAAUACUA ---.((((((...((.........---))...)------))))).(((...((((((((((((..(((((((............)))))))...------...-)))))))))))).))) ( -20.70) >DroYak_CAF1 2762 90 - 1 --------------GUUUUGUAUU---UACCAU------UCGCACUAGGCGGUUUCUACAAUUUCACUUGACAUUAAUGUCAAAGUGAAGUUUA------GUC-GGUUGUGGAAAUACUA --------------.....(((((---(.((((------(((.(((((((.((....))...(((((((((((....)))).))))))))))))------)))-))..)))))))))).. ( -28.10) >DroAna_CAF1 2301 99 - 1 ---------------UUCGGGUUUCUACCCCAU------UCGUACUAGGCCACUUUCUCAAUUUCAUUUGACAUUAAUGUCAAAGUCAAAAUCAAAGUCAUUUGGUUUGUGGAAAUCCUA ---------------...((((((((((.....------...........................(((((((....)))))))....((((((((....)))))))))))))))))).. ( -23.90) >consensus ____UAUGAAACUGAUACGGAUUU___UCCCAU______UCGUACUAGGCGGUUUCUACAAUUUCACUUGACAAUAAUGUCAAAGUCAAGUUCA______GUC_GGUUGUGGAAAUGCUA ........................................(((.....)))((((((((((((..(((((((............))))))).............)))))))))))).... (-13.44 = -13.56 + 0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:05 2006