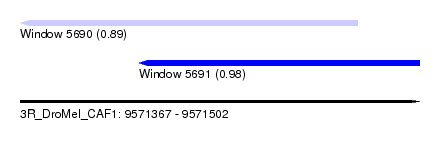
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,571,367 – 9,571,502 |
| Length | 135 |
| Max. P | 0.979790 |

| Location | 9,571,367 – 9,571,481 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.84 |
| Mean single sequence MFE | -50.43 |
| Consensus MFE | -35.53 |
| Energy contribution | -34.23 |
| Covariance contribution | -1.30 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9571367 114 - 27905053 UCGUCUGGUAGGCCAGCGGCAACUUCUUCUGCUGCUGCGCCAGGAUCGCUGCCUCCUG------CAGCUGCUGCAGCUUCUUGGCGUCCAUGGUGCCGGCGGUGAAGAGGGGCGGCGCCU .........((((..((.((..(((((((.(((((((((((((((.((((.....(((------(((...))))))......))))))).))))).)))))))))))))).)).)))))) ( -54.70) >DroVir_CAF1 62425 111 - 1 UUGUUUGAUAGGCCAGCGGCAGUUUCUUCUGCUGCUGGGCUG---CCGUUGCCGCCUG------UAACUGUUGCAGCUUCUUGGCGUCCAUGGUGCCGGCCGUGAAGAGCGGUGGCGCCU ........(((.((((((((((......)))))))))).)))---.(((..(((((((------((.....)))))..((((.(((.((........)).))).))))))))..)))... ( -51.70) >DroGri_CAF1 59614 108 - 1 UCGUCUGAUAGGCGAGUGGCAGUUUCUUUUGCUGCUGUG------CAGUUGCCGCCUG------CAACUGUUGCAGCUUUUUGGCGUCCAUGGUGCCGGCGGUGAAGAGCGGUGGCGCUU (((((.....)))))((((..(((......(((((...(------(((((((.....)------))))))).))))).....)))..))))(((((((.((.(....).)).))))))). ( -44.80) >DroWil_CAF1 53906 114 - 1 UAGUUUGAUAGGCCAGCGGCAGCUUUUUCUGUUGCUGGGCCAAAGCAGCUGCCUCUUG------CAACUGUUGCAGCUUUUUGGGGUCCAUAGUGCCAGCUGUGAAAAGUGGUGGAGCUU .(((((.((...((((((((((......)))))))))).((((((.((((((..(...------.....)..))))))))))))....((((((....)))))).......)).))))). ( -41.60) >DroMoj_CAF1 56823 114 - 1 UGGUUUGAUAGGCCAGCGGCAGCUUCUUCUGUUGCUGGG------CCGUUGCCGCCUGCAGCUGCAACUGCUGCAGCUUCUUUGGGUCCAUGGUGCCGCCUGUGAAGAGCGGUGGCGCCU (((((..(((((((((((((((......)))))))))((------(....)))))))).(((((((.....)))))))...)..)).))).(((((((((.((.....))))))))))). ( -55.00) >DroPer_CAF1 46599 114 - 1 UCGUUUGGUAGGCCAGCGGCAGCUUUUUCUGCUGCUGGGCGAGUAUGGCCGCCUCCUG------CAGCUGCUGCAACUUCUUGGCGUCCAUGGUGCCGGCCGUGAAGAGCGGUGGCGCCU ..........(.((((((((((......)))))))))).)....((((.((((...((------(((...))))).......)))).))))(((((((.((((.....))))))))))). ( -54.80) >consensus UCGUUUGAUAGGCCAGCGGCAGCUUCUUCUGCUGCUGGGC_A___CCGCUGCCGCCUG______CAACUGCUGCAGCUUCUUGGCGUCCAUGGUGCCGGCCGUGAAGAGCGGUGGCGCCU .........(((((((((((((......)))))))))..........((((((((((.........((.((((..(((...(((...))).)))..)))).))..)).)))))))))))) (-35.53 = -34.23 + -1.30)



| Location | 9,571,407 – 9,571,502 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 75.97 |
| Mean single sequence MFE | -41.97 |
| Consensus MFE | -27.70 |
| Energy contribution | -27.40 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9571407 95 - 27905053 -------------------CCGUUCGGUAGUCCACGAGAUUCGUCUGGUAGGCCAGCGGCAACUUCUUCUGCUGCUGCGCCAGGAUCGCUGCCUCCUGCAGCUGCUGCAGCUUC -------------------..((((((((((((((.(((....))).)).(((((((((((........)))))))).))).)))).(((((.....)))))))))).)))... ( -40.80) >DroVir_CAF1 62465 105 - 1 G------GCGAAUGUGUUGCCGUGCGAUAAUCAAUCAGAUUUGUUUGAUAGGCCAGCGGCAGUUUCUUCUGCUGCUGGGCUG---CCGUUGCCGCCUGUAACUGUUGCAGCUUC (------((((.....))))).(((((((....((((((....))))))(((((((((((((......)))))))))(((..---.....))))))).....)))))))..... ( -42.30) >DroPse_CAF1 46538 108 - 1 G------GCGAAUGUGUGGCCGUGCGGUAGUCCAUGAGAUUCGUUUGGUACGCCAGCGGCAGCUUUUUCUGCUGCUGGGCCAGUAUGGCCGCCUCCUGCAGCUGCUGCAACUUC (------(((..((((((((((((((((.(((((...........))).)).((((((((((......))))))))))))).)))))))))).....)))..))))........ ( -47.60) >DroWil_CAF1 53946 108 - 1 ACUGCCGACGAUUG------UGUGCGAUAGUCAAUGAGAUUAGUUUGAUAGGCCAGCGGCAGCUUUUUCUGUUGCUGGGCCAAAGCAGCUGCCUCUUGCAACUGUUGCAGCUUU .((((.(((.((((------....)))).)))..................((((((((((((......)))))))).))))..(((((.(((.....))).))))))))).... ( -37.00) >DroAna_CAF1 56151 95 - 1 -------------------CCGUCCGGUAGUCCACCAGAUUGGUUUGGUACGCCAGCGGCAGCUUUUUCUGCUGCUGCGCCAGUAUGGCCGCCUCCUGCAGCUGCUGCAGCUUC -------------------......((..(.(((....((((((.(((....)))((((((((.......)))))))))))))).))))..))..((((((...)))))).... ( -38.50) >DroPer_CAF1 46639 108 - 1 G------GCGAAUGUGUGGCCGUGCGGUAGUCCAUGAGAUUCGUUUGGUAGGCCAGCGGCAGCUUUUUCUGCUGCUGGGCGAGUAUGGCCGCCUCCUGCAGCUGCUGCAACUUC (------(((..(((((((((((((((....)).................(.((((((((((......)))))))))).)..)))))))))).....)))..))))........ ( -45.60) >consensus G______GCGAAUG_____CCGUGCGGUAGUCCAUGAGAUUCGUUUGGUAGGCCAGCGGCAGCUUUUUCUGCUGCUGGGCCAGUAUGGCCGCCUCCUGCAGCUGCUGCAGCUUC ......................(((((((((..((.(((....))).)).((((((((((((......)))))))))(((.......)))))).......)))))))))..... (-27.70 = -27.40 + -0.30)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:45 2006