
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,557,472 – 9,557,607 |
| Length | 135 |
| Max. P | 0.974473 |

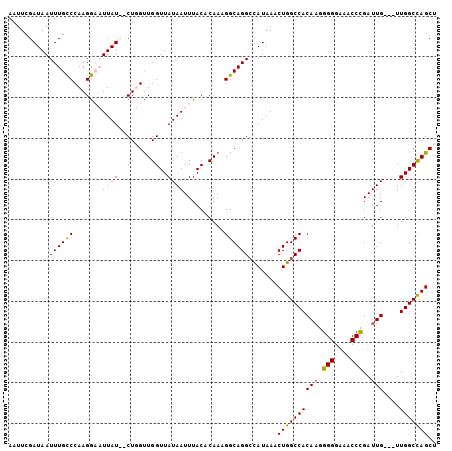
| Location | 9,557,472 – 9,557,575 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 88.85 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -23.15 |
| Energy contribution | -23.65 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9557472 103 + 27905053 AAUUCGAUAAUUUGCCCAAGGCAUUAU--CUUGUUGGUUAUAAUUUACCCAAAGGCAGGCCAUAAACUGGCCACAAGGGGGAAACCCGAUUGUUGUUGGCCAGCU .....(((((..((((...))))))))--)..((((((((.................(((((.....)))))((((.(((....)))..))))...)))))))). ( -34.50) >DroSec_CAF1 34124 100 + 1 AAUUCGAUAAUUUGCCCAAGGCAUUAU--CUUGUUGGUUAUAAUUUACACAAAGGCAGGCCAUAAACUGGCCACAUAGGGGAAACCUGAUUG---UUGGCCAGCU .....(((((..((((...))))))))--)..((((((((.................(((((.....)))))(((..(((....)))...))---))))))))). ( -32.10) >DroEre_CAF1 35781 100 + 1 AAUUCGAUAAUUUGUCCAAGAA-UUAUCUCUGGUUGGUUAUAAUUUACACAACGGCAGGACAUAAACUGGCCACAAGGGGGA-ACCCGAUUG---UUGGCUAGCU ............(((((.(((.-.....))).((((((........)).))))....)))))....(((((((((.(((...-.)))...))---.))))))).. ( -29.50) >DroYak_CAF1 34396 102 + 1 AAUUCGAUAAUUUGCCCAAGGAUUUAUUUCUGGCUGGUUAUAAUUUACUCAAAGGCAGGCUAUAAACUGGCCACAAGGGGGAAACCCGAUUG---UUGGCCAGCU ......(((((..(((..((((.....)))))))..))))).........................((((((((((.(((....)))..)))---.))))))).. ( -32.30) >consensus AAUUCGAUAAUUUGCCCAAGGAAUUAU__CUGGUUGGUUAUAAUUUACACAAAGGCAGGCCAUAAACUGGCCACAAGGGGGAAACCCGAUUG___UUGGCCAGCU ..........((((((....(((((((..((....))..))))))).......)))))).......((((((((((.(((....)))..)))....))))))).. (-23.15 = -23.65 + 0.50)


| Location | 9,557,472 – 9,557,575 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 88.85 |
| Mean single sequence MFE | -26.24 |
| Consensus MFE | -17.71 |
| Energy contribution | -18.40 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9557472 103 - 27905053 AGCUGGCCAACAACAAUCGGGUUUCCCCCUUGUGGCCAGUUUAUGGCCUGCCUUUGGGUAAAUUAUAACCAACAAG--AUAAUGCCUUGGGCAAAUUAUCGAAUU .((((((((.(((.....(((.....))))))))))))))((((((..((((....))))..)))))).......(--((((((((...))))..)))))..... ( -30.10) >DroSec_CAF1 34124 100 - 1 AGCUGGCCAA---CAAUCAGGUUUCCCCUAUGUGGCCAGUUUAUGGCCUGCCUUUGUGUAAAUUAUAACCAACAAG--AUAAUGCCUUGGGCAAAUUAUCGAAUU (((((((((.---..((.(((.....))))).)))))))))...((....)).((((..............))))(--((((((((...))))..)))))..... ( -24.94) >DroEre_CAF1 35781 100 - 1 AGCUAGCCAA---CAAUCGGGU-UCCCCCUUGUGGCCAGUUUAUGUCCUGCCGUUGUGUAAAUUAUAACCAACCAGAGAUAA-UUCUUGGACAAAUUAUCGAAUU ((((.((((.---(((..(((.-...)))))))))).))))..(((((....((((((.....))))))......((((...-.)))))))))............ ( -24.70) >DroYak_CAF1 34396 102 - 1 AGCUGGCCAA---CAAUCGGGUUUCCCCCUUGUGGCCAGUUUAUAGCCUGCCUUUGAGUAAAUUAUAACCAGCCAGAAAUAAAUCCUUGGGCAAAUUAUCGAAUU .((((((((.---(((..(((....))).)))))))))))((((((..(((......)))..))))))...(((.((......))....)))............. ( -25.20) >consensus AGCUGGCCAA___CAAUCGGGUUUCCCCCUUGUGGCCAGUUUAUGGCCUGCCUUUGUGUAAAUUAUAACCAACAAG__AUAAUGCCUUGGGCAAAUUAUCGAAUU (((((((((....(((..(((.....))))))))))))))).......((((.(((.((........)))))((((.........))))))))............ (-17.71 = -18.40 + 0.69)



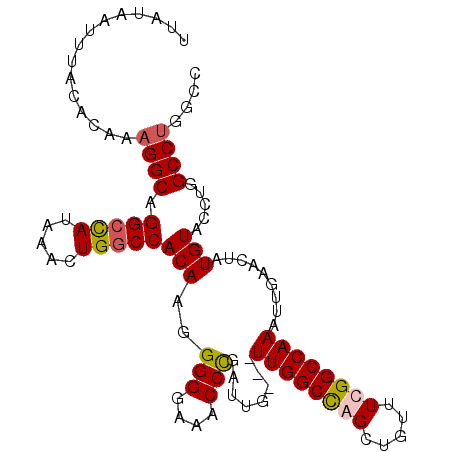
| Location | 9,557,507 – 9,557,607 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 89.92 |
| Mean single sequence MFE | -35.02 |
| Consensus MFE | -28.06 |
| Energy contribution | -28.50 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9557507 100 + 27905053 UUAUAAUUUACCCAAAGGCAGGCCAUAAACUGGCCACAAGGGGGAAACCCGAUUGUUGUUGGCCAGCUGUUUUGGCCAAAUUGA----UGUACCUUGCCUGGCC ...........(((..(((((((((.....)))))((((.(((....)))..))))..((((((((.....)))))))).....----.......))))))).. ( -36.50) >DroSec_CAF1 34159 101 + 1 UUAUAAUUUACACAAAGGCAGGCCAUAAACUGGCCACAUAGGGGAAACCUGAUUG---UUGGCCAGCUGUUUUGGCCAAAUUGAACUAUGUACCUGGCCUGGCC ...............((((.(((((.....)))))((((((((....))......---((((((((.....))))))))......)))))).....)))).... ( -36.70) >DroEre_CAF1 35817 100 + 1 UUAUAAUUUACACAACGGCAGGACAUAAACUGGCCACAAGGGGGA-ACCCGAUUG---UUGGCUAGCUGUUUCGGCCAAAUUGAACUAUGUACCUGGCCUGGCU ................(((((((((((..(((((((((.(((...-.)))...))---.)))))))....(((((.....))))).))))).))).)))..... ( -30.90) >DroYak_CAF1 34433 101 + 1 UUAUAAUUUACUCAAAGGCAGGCUAUAAACUGGCCACAAGGGGGAAACCCGAUUG---UUGGCCAGCUGUUUCGGCCAAAUUGAACUAUGUACCUUGCCUGGCU ...............(((((((.((((..((((((((((.(((....)))..)))---.)))))))....(((((.....)))))...)))).))))))).... ( -36.00) >consensus UUAUAAUUUACACAAAGGCAGGCCAUAAACUGGCCACAAGGGGGAAACCCGAUUG___UUGGCCAGCUGUUUCGGCCAAAUUGAACUAUGUACCUGGCCUGGCC ...............((((.(((((.....)))))(((..(((....)))........((((((((.....)))))))).........))).....)))).... (-28.06 = -28.50 + 0.44)


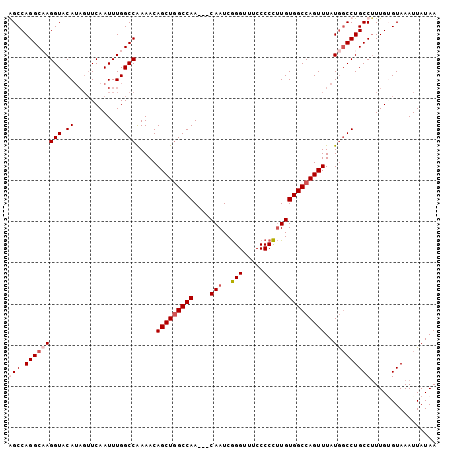
| Location | 9,557,507 – 9,557,607 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 89.92 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -24.88 |
| Energy contribution | -26.25 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9557507 100 - 27905053 GGCCAGGCAAGGUACA----UCAAUUUGGCCAAAACAGCUGGCCAACAACAAUCGGGUUUCCCCCUUGUGGCCAGUUUAUGGCCUGCCUUUGGGUAAAUUAUAA ((((((((.((((...----...)))).))).....(((((((((.(((.....(((.....)))))))))))))))..)))))((((....))))........ ( -36.00) >DroSec_CAF1 34159 101 - 1 GGCCAGGCCAGGUACAUAGUUCAAUUUGGCCAAAACAGCUGGCCAA---CAAUCAGGUUUCCCCUAUGUGGCCAGUUUAUGGCCUGCCUUUGUGUAAAUUAUAA (((((((((((((..........)))))))).....(((((((((.---..((.(((.....))))).)))))))))..))))).................... ( -36.60) >DroEre_CAF1 35817 100 - 1 AGCCAGGCCAGGUACAUAGUUCAAUUUGGCCGAAACAGCUAGCCAA---CAAUCGGGU-UCCCCCUUGUGGCCAGUUUAUGUCCUGCCGUUGUGUAAAUUAUAA .....(((.(((.((((((......(((((((........((((..---......)))-)........))))))).))))))))))))................ ( -26.49) >DroYak_CAF1 34433 101 - 1 AGCCAGGCAAGGUACAUAGUUCAAUUUGGCCGAAACAGCUGGCCAA---CAAUCGGGUUUCCCCCUUGUGGCCAGUUUAUAGCCUGCCUUUGAGUAAAUUAUAA ((.(((((..(((.((..........))))).....(((((((((.---(((..(((....))).))))))))))))....))))).))............... ( -31.60) >consensus AGCCAGGCAAGGUACAUAGUUCAAUUUGGCCAAAACAGCUGGCCAA___CAAUCGGGUUUCCCCCUUGUGGCCAGUUUAUGGCCUGCCUUUGUGUAAAUUAUAA (((.(((((((((.((..........))))).....(((((((((....(((..(((.....)))))))))))))))..)))))))))................ (-24.88 = -26.25 + 1.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:40 2006