
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,551,615 – 9,551,740 |
| Length | 125 |
| Max. P | 0.979465 |

| Location | 9,551,615 – 9,551,715 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 85.96 |
| Mean single sequence MFE | -31.75 |
| Consensus MFE | -22.65 |
| Energy contribution | -23.99 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9551615 100 + 27905053 CUCACUUUUCGCUGCAGUUCAGUGGCCGUUGCACUUGACAUAUAGCCACUG-UCAAUGCGACUAAGAGUGCAACUGGAAUGGCCAUUGAAAUUGCAACUGC ............(((((((((((((((((((((.((((((.........))-))))))))))....((.....)).....)))))))).)))))))..... ( -34.60) >DroSec_CAF1 28240 100 + 1 CUCACUUUUCGCUGCAGUUCAGUGGCCGUUGCACUUGACAUACAGUCACUG-UCAAUGCGACUAAAAGUGCAACUGGAAUAGCCAUUGAAAUUGCAACUGC ............((((((((((((((.((((((((((((.....)))...(-((.....)))...))))))))).......))))))).)))))))..... ( -32.80) >DroSim_CAF1 28381 100 + 1 CUCACUUUUCGCUGCAGUUCAGUGGCCGUUGCACUUGACAUACAGCCACUG-UCAAUGCGACUAAAAGUGCAACUGGAAUGGUCAUUGAAAUUGCAACUGC ............((((((((((((((((((.((.((((((.........))-))))(((.((.....)))))..)).))))))))))).)))))))..... ( -31.80) >DroEre_CAF1 29984 100 + 1 CUCACUUUUCGCUGCAGUUCAGUGGCCGUUGCACUUGACAUAUAGCCACUG-UCAAUGCGACUAAAAGUGCAACUGGAAUGGCCAUUGAAAUUGCAACUGC ............((((((((((((((((((.((.((((((.........))-))))(((.((.....)))))..)).))))))))))).)))))))..... ( -34.50) >DroWil_CAF1 32410 96 + 1 ACCAUUAUUUGAUGCGGUUCAGUGGCCAAUGCACUUGACACUCUGCCACUGAUUGUUGCAACUGAAACUGGAACUGGAAUUGGAAUUGGCCUUUCA----- .(((......(.(((((.((((((((...(((....).))....))))))))...))))).)......))).....(((..((......)).))).----- ( -21.50) >DroYak_CAF1 28665 95 + 1 CUCACUUUUCG---CAGUUCAGUGGCCGUUGCACUUGACAUGUAGCCACUG-UCAAUGCGACUAAAAGUGCAACUGGAAUGGCCAUUGAAAUUGCAACU-- ..........(---((((((((((((((((.((.((((((.((....))))-))))(((.((.....)))))..)).))))))))))).))))))....-- ( -35.30) >consensus CUCACUUUUCGCUGCAGUUCAGUGGCCGUUGCACUUGACAUACAGCCACUG_UCAAUGCGACUAAAAGUGCAACUGGAAUGGCCAUUGAAAUUGCAACUGC ............((((((((((((((((((((((((........((...........))......)))))))))......))))))))).))))))..... (-22.65 = -23.99 + 1.34)



| Location | 9,551,615 – 9,551,715 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 85.96 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -24.75 |
| Energy contribution | -26.83 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9551615 100 - 27905053 GCAGUUGCAAUUUCAAUGGCCAUUCCAGUUGCACUCUUAGUCGCAUUGA-CAGUGGCUAUAUGUCAAGUGCAACGGCCACUGAACUGCAGCGAAAAGUGAG ...((((((..((((.(((((......((((((((..(((((((.....-..))))))).......))))))))))))).)))).)))))).......... ( -36.60) >DroSec_CAF1 28240 100 - 1 GCAGUUGCAAUUUCAAUGGCUAUUCCAGUUGCACUUUUAGUCGCAUUGA-CAGUGACUGUAUGUCAAGUGCAACGGCCACUGAACUGCAGCGAAAAGUGAG ...((((((..((((.(((((......(((((((((.(((((((.....-..)))))))......)))))))))))))).)))).)))))).......... ( -36.30) >DroSim_CAF1 28381 100 - 1 GCAGUUGCAAUUUCAAUGACCAUUCCAGUUGCACUUUUAGUCGCAUUGA-CAGUGGCUGUAUGUCAAGUGCAACGGCCACUGAACUGCAGCGAAAAGUGAG ((((((((((((..(((....)))..)))))).................-((((((((((.(((.....)))))))))))))))))))............. ( -31.90) >DroEre_CAF1 29984 100 - 1 GCAGUUGCAAUUUCAAUGGCCAUUCCAGUUGCACUUUUAGUCGCAUUGA-CAGUGGCUAUAUGUCAAGUGCAACGGCCACUGAACUGCAGCGAAAAGUGAG ...((((((..((((.(((((......(((((((((.(((((((.....-..)))))))......)))))))))))))).)))).)))))).......... ( -37.70) >DroWil_CAF1 32410 96 - 1 -----UGAAAGGCCAAUUCCAAUUCCAGUUCCAGUUUCAGUUGCAACAAUCAGUGGCAGAGUGUCAAGUGCAUUGGCCACUGAACCGCAUCAAAUAAUGGU -----......((((............(((.(((......))).)))..((((((((..(((((.....))))).))))))))..............)))) ( -23.00) >DroYak_CAF1 28665 95 - 1 --AGUUGCAAUUUCAAUGGCCAUUCCAGUUGCACUUUUAGUCGCAUUGA-CAGUGGCUACAUGUCAAGUGCAACGGCCACUGAACUG---CGAAAAGUGAG --..(((((..((((.(((((......(((((((((.(((((((.....-..)))))))......)))))))))))))).)))).))---)))........ ( -33.10) >consensus GCAGUUGCAAUUUCAAUGGCCAUUCCAGUUGCACUUUUAGUCGCAUUGA_CAGUGGCUAUAUGUCAAGUGCAACGGCCACUGAACUGCAGCGAAAAGUGAG ...((((((..((((.(((((......(((((((((.(((((((........)))))))......)))))))))))))).)))).)))))).......... (-24.75 = -26.83 + 2.09)

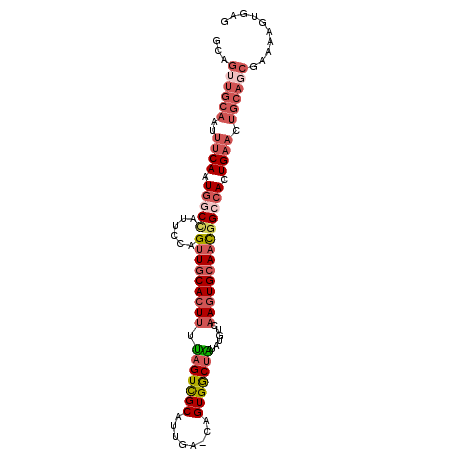

| Location | 9,551,642 – 9,551,740 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 78.48 |
| Mean single sequence MFE | -25.15 |
| Consensus MFE | -13.70 |
| Energy contribution | -15.07 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.803916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9551642 98 + 27905053 GUUGCACUUGACAUAUAGCCACUG-UCAAUGCGACUAAGAGUGCAACUGGAAUGGCCAUUGAAAUUGCAACUGCAAUUGCUAUUGCAACUGCAACUGCC ((((((.((((((.........))-))))))))))....((((((..((.((((((......((((((....)))))))))))).))..))).)))... ( -30.00) >DroSec_CAF1 28267 98 + 1 GUUGCACUUGACAUACAGUCACUG-UCAAUGCGACUAAAAGUGCAACUGGAAUAGCCAUUGAAAUUGCAACUGCAAUUGCAAUUGCAACUGCAACUGCC ((((((((((((.....)))...(-((.....)))...)))))))))(((.....)))....((((((....))))))(((.((((....)))).))). ( -30.50) >DroSim_CAF1 28408 98 + 1 GUUGCACUUGACAUACAGCCACUG-UCAAUGCGACUAAAAGUGCAACUGGAAUGGUCAUUGAAAUUGCAACUGCAAUUGCAAUUGCAACUGCAACUGCC ((((((..((((...(((.(((((-((.....)))....))))...))).....))))......((((((.(((....))).)))))).)))))).... ( -27.30) >DroEre_CAF1 30011 98 + 1 GUUGCACUUGACAUAUAGCCACUG-UCAAUGCGACUAAAAGUGCAACUGGAAUGGCCAUUGAAAUUGCAACUGCAAUUGCAAGUGCAACUGCAACUACC ((((((((((.(.....((((...-(((.(((.((.....)))))..)))..))))......((((((....))))))))))))))))).......... ( -30.20) >DroWil_CAF1 32437 80 + 1 AAUGCACUUGACACUCUGCCACUGAUUGUUGCAACUGAAACUGGAACUGGAAUUGGAAUUGGCCUUUCA-------------------UUGUGAAUGCC ...(((.((.(((....(((((..((((((.((........)).)))...)))..)...))))......-------------------.))).))))). ( -12.40) >DroYak_CAF1 28689 80 + 1 GUUGCACUUGACAUGUAGCCACUG-UCAAUGCGACUAAAAGUGCAACUGGAAUGGCCAUUGAAAUUGCAACU------------------GCAACUGCC ((((((.((((((.((....))))-))))))))))....((((((..((.(((..(....)..))).))..)------------------)).)))... ( -20.50) >consensus GUUGCACUUGACAUACAGCCACUG_UCAAUGCGACUAAAAGUGCAACUGGAAUGGCCAUUGAAAUUGCAACUGCAAUUGCAAUUGCAACUGCAACUGCC (((((((((........((...........))......))))))))).................(((((..((((((....))))))..)))))..... (-13.70 = -15.07 + 1.38)



| Location | 9,551,642 – 9,551,740 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 78.48 |
| Mean single sequence MFE | -32.78 |
| Consensus MFE | -24.46 |
| Energy contribution | -26.43 |
| Covariance contribution | 1.97 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.73 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9551642 98 - 27905053 GGCAGUUGCAGUUGCAAUAGCAAUUGCAGUUGCAAUUUCAAUGGCCAUUCCAGUUGCACUCUUAGUCGCAUUGA-CAGUGGCUAUAUGUCAAGUGCAAC (((.((((.(((((((((.((....)).))))))))).)))).)))......((((((((..(((((((.....-..))))))).......)))))))) ( -38.40) >DroSec_CAF1 28267 98 - 1 GGCAGUUGCAGUUGCAAUUGCAAUUGCAGUUGCAAUUUCAAUGGCUAUUCCAGUUGCACUUUUAGUCGCAUUGA-CAGUGACUGUAUGUCAAGUGCAAC (((.((((.((((((((((((....)))))))))))).)))).)))......(((((((((.(((((((.....-..)))))))......))))))))) ( -40.90) >DroSim_CAF1 28408 98 - 1 GGCAGUUGCAGUUGCAAUUGCAAUUGCAGUUGCAAUUUCAAUGACCAUUCCAGUUGCACUUUUAGUCGCAUUGA-CAGUGGCUGUAUGUCAAGUGCAAC ((..((((.((((((((((((....)))))))))))).))))..))......(((((((((.(((((((.....-..)))))))......))))))))) ( -39.90) >DroEre_CAF1 30011 98 - 1 GGUAGUUGCAGUUGCACUUGCAAUUGCAGUUGCAAUUUCAAUGGCCAUUCCAGUUGCACUUUUAGUCGCAUUGA-CAGUGGCUAUAUGUCAAGUGCAAC (((.((((.(((((((.((((....)))).))))))).)))).)))......(((((((((.(((((((.....-..)))))))......))))))))) ( -37.80) >DroWil_CAF1 32437 80 - 1 GGCAUUCACAA-------------------UGAAAGGCCAAUUCCAAUUCCAGUUCCAGUUUCAGUUGCAACAAUCAGUGGCAGAGUGUCAAGUGCAUU (((((((....-------------------(((((((..((((........))))))..)))))((..(........)..)).)))))))......... ( -14.70) >DroYak_CAF1 28689 80 - 1 GGCAGUUGC------------------AGUUGCAAUUUCAAUGGCCAUUCCAGUUGCACUUUUAGUCGCAUUGA-CAGUGGCUACAUGUCAAGUGCAAC (((((((((------------------....))))))......)))......(((((((((.(((((((.....-..)))))))......))))))))) ( -25.00) >consensus GGCAGUUGCAGUUGCAAUUGCAAUUGCAGUUGCAAUUUCAAUGGCCAUUCCAGUUGCACUUUUAGUCGCAUUGA_CAGUGGCUAUAUGUCAAGUGCAAC .((((((((((((((....))))))))))))))...................(((((((((.(((((((........)))))))......))))))))) (-24.46 = -26.43 + 1.97)

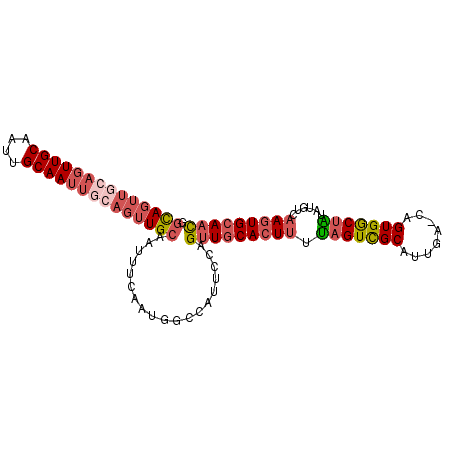
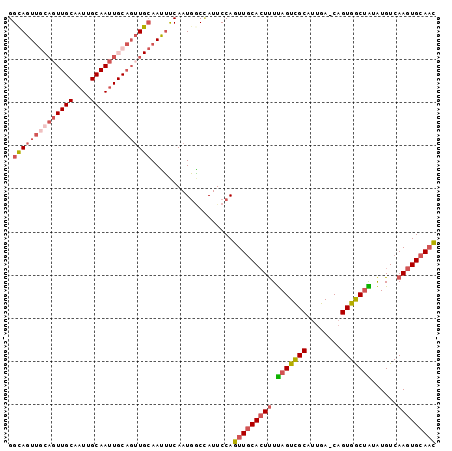
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:35 2006