
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,525,992 – 9,526,083 |
| Length | 91 |
| Max. P | 0.707152 |

| Location | 9,525,992 – 9,526,083 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 75.32 |
| Mean single sequence MFE | -19.33 |
| Consensus MFE | -13.90 |
| Energy contribution | -13.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9525992 91 + 27905053 -CGUACAGUG---------UACCCAGCCACCACCCACCCACAACCCUCGGAAAAUGUUUGUUUUCCGAAGCGGCAAAAAUGCUGAGCC-----ACUUGUCUUUGCU -.(.((((((---------...........................(((((((((....)))))))))..(((((....)))))...)-----)).))))...... ( -18.40) >DroPse_CAF1 2857 87 + 1 -UUUGCAG------------------CCAAUGCACACCUACGCCACUCGCCUCAAGCGAGUCCGACGAAGCGGCAAAAAUGCUGAGCCCCGGCAUUUGCCUUCCCC -..((((.------------------....))))......((..((((((.....)))))).)).......(((((..((((((.....)))))))))))...... ( -25.30) >DroSec_CAF1 2999 88 + 1 GUGUACAGUG---------UACCCA-C---CACCCACCCACAACCCUCGGAAAAUGUUUGUUUUCCGAAGCGGCAAAAAUGCUGAGCC-----ACUUGUCUUUGCU (((.((((((---------......-.---................(((((((((....)))))))))..(((((....)))))...)-----)).)))...))). ( -18.30) >DroSim_CAF1 3000 91 + 1 GUGUACAGUG---------UACCCA-CCACCACCCACCCACAACCCUCGGAAAAUGUUUGUUUUCCGAAGCGGCAAAAAUGCUGAGCC-----ACUUGUCUUUGCU (((.((((((---------......-....................(((((((((....)))))))))..(((((....)))))...)-----)).)))...))). ( -18.30) >DroEre_CAF1 3106 81 + 1 -CAUACAGU-------------------ACCCACCACCCACAAACCUCGGAAAAUGUUUAUUUUCCGAAGCGGCAAAAAUGCUGAGCC-----ACUUGUCUUUGCU -.....(((-------------------(.................(((((((((....)))))))))..(((((....)))))....-----.........)))) ( -15.30) >DroYak_CAF1 3144 99 + 1 -CCUACAGUACAUGCCCUCUACGCA-CCACCCACCACCCACAACCCUCGGAAAAUGUUUGUUUUCCGAAGCGGCAAAAAUGCUGAGCC-----ACUUGUAUUUGCU -.....((((.((((.......((.-....................(((((((((....)))))))))..(((((....))))).)).-----....)))).)))) ( -20.40) >consensus _CGUACAGUG_________UACCCA_CCACCACCCACCCACAACCCUCGGAAAAUGUUUGUUUUCCGAAGCGGCAAAAAUGCUGAGCC_____ACUUGUCUUUGCU ....((((......................................(((((((((....)))))))))..(((((....)))))...........))))....... (-13.90 = -13.90 + 0.00)
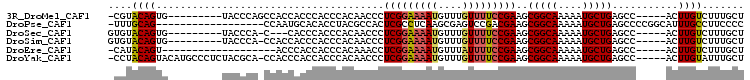
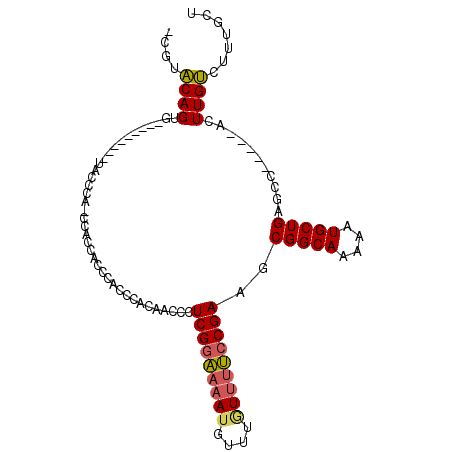

| Location | 9,525,992 – 9,526,083 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 75.32 |
| Mean single sequence MFE | -29.75 |
| Consensus MFE | -16.60 |
| Energy contribution | -16.72 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9525992 91 - 27905053 AGCAAAGACAAGU-----GGCUCAGCAUUUUUGCCGCUUCGGAAAACAAACAUUUUCCGAGGGUUGUGGGUGGGUGGUGGCUGGGUA---------CACUGUACG- .......(((.((-----((((((((...((..((.((((((((((......))))))))))......))..)).....))))))).---------))))))...- ( -30.30) >DroPse_CAF1 2857 87 - 1 GGGGAAGGCAAAUGCCGGGGCUCAGCAUUUUUGCCGCUUCGUCGGACUCGCUUGAGGCGAGUGGCGUAGGUGUGCAUUGG------------------CUGCAAA- .(((..(((....)))....))).(((((..((((((((((((..((((((.....)))))))))).))))).)))..))------------------.)))...- ( -34.10) >DroSec_CAF1 2999 88 - 1 AGCAAAGACAAGU-----GGCUCAGCAUUUUUGCCGCUUCGGAAAACAAACAUUUUCCGAGGGUUGUGGGUGGGUG---G-UGGGUA---------CACUGUACAC .......(((.((-----((((((.(((((...(((((((((((((......)))))))))....))))..)))))---.-))))).---------)))))).... ( -28.20) >DroSim_CAF1 3000 91 - 1 AGCAAAGACAAGU-----GGCUCAGCAUUUUUGCCGCUUCGGAAAACAAACAUUUUCCGAGGGUUGUGGGUGGGUGGUGG-UGGGUA---------CACUGUACAC .......(((.((-----((((((.((((((..((.((((((((((......))))))))))......))..)).)))).-))))).---------)))))).... ( -28.70) >DroEre_CAF1 3106 81 - 1 AGCAAAGACAAGU-----GGCUCAGCAUUUUUGCCGCUUCGGAAAAUAAACAUUUUCCGAGGUUUGUGGGUGGUGGGU-------------------ACUGUAUG- .......(((.((-----(.((((.(((((.....((((((((((((....))))))))))))....))))).)))))-------------------)))))...- ( -26.90) >DroYak_CAF1 3144 99 - 1 AGCAAAUACAAGU-----GGCUCAGCAUUUUUGCCGCUUCGGAAAACAAACAUUUUCCGAGGGUUGUGGGUGGUGGGUGG-UGCGUAGAGGGCAUGUACUGUAGG- ......((((.((-----((((((.(((((..(((.((.(((((((......))))))))))))...))))).))))).(-(((.......)))).)))))))..- ( -30.30) >consensus AGCAAAGACAAGU_____GGCUCAGCAUUUUUGCCGCUUCGGAAAACAAACAUUUUCCGAGGGUUGUGGGUGGGUGGUGG_UGGGUA_________CACUGUACA_ ...................((((((((....)))..((((((((((......))))))))))....)))))................................... (-16.60 = -16.72 + 0.11)

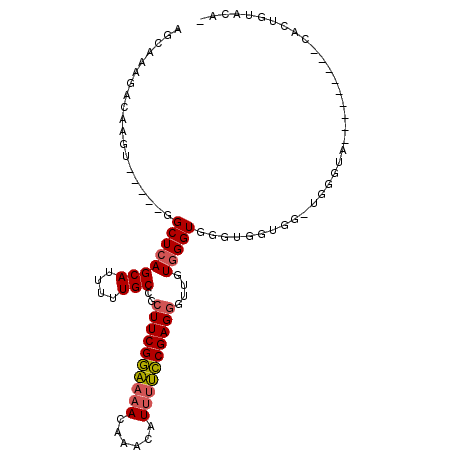
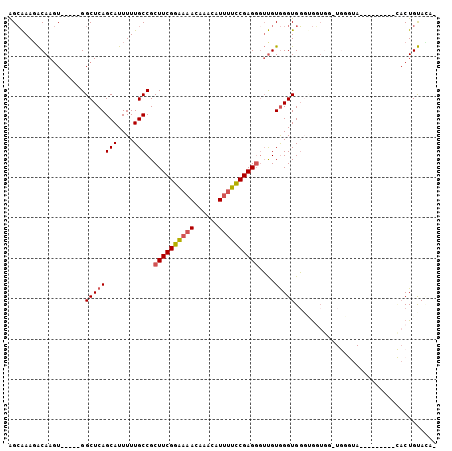
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:25 2006