| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 9,510,839 – 9,511,199 |
| Length | 360 |
| Max. P | 0.996957 |

| Location | 9,510,839 – 9,510,959 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.39 |
| Mean single sequence MFE | -39.24 |
| Consensus MFE | -32.96 |
| Energy contribution | -32.96 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
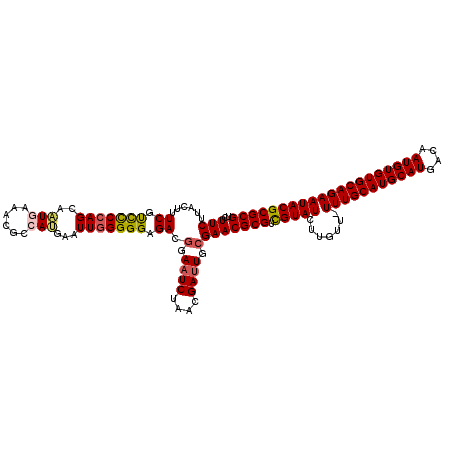
>3R_DroMel_CAF1 9510839 120 - 27905053 UGUUUUUUGCAUGCAUGACAAUGUGUGCAGAAUACGCGCGCGCUCACACACUAAACCACUCGCACAACCGAAGGCGUGUGUGCAUGUAUGCAAUUUUUUGGGGUGGUAUUGGCGGUGAAG .((.((((((((((((....)))))))))))).))((....))((((.(.((((((((((((((((..((....))..))))).......(((....)))))))))).)))).))))).. ( -44.50) >DroSec_CAF1 140517 119 - 1 UGUU-UUUGCAUGCAUGACAAUGUGUGCAGAAUACGCGCGCGCUCACACACUAAACCUCUCGCACAACCGAAGGCGUGUGUGCAUGUAUGCAAUUUUUUGGGGUGGUAUUGGCGGUGAAG .(((-(((((((((((....)))))))))))).))((....))((((.(.(((((((.((((((((..((....))..))))).......(((....)))))).))).)))).))))).. ( -39.10) >DroSim_CAF1 130678 119 - 1 UGUU-UUUGCAUGCAUGACAAUGUGUGCAGAAUACGCGCGCGCUCACACACUAAACCUCUCGCACAACCGAAGGCGUGUGUGCAUGUAUGCAAUUUUUUGGGGUGGUAUUGGCGGUGAAG .(((-(((((((((((....)))))))))))).))((....))((((.(.(((((((.((((((((..((....))..))))).......(((....)))))).))).)))).))))).. ( -39.10) >DroEre_CAF1 148369 116 - 1 UCUU-UUUGCAUGCAUGACAAUGUGUGCAGAAUACGCGCGCGCUCACACACUAAAACAUUCGCACAACCGAAGGCGUGUGUGCAUGUAUGCAAUUUUUUGGGGUGGUAUUGGCGCAG--- ...(-(((((((((((....)))))))))))).......(((((.((.((((...((((.((((((..(....)..)))))).))))...(((....))).))))))...)))))..--- ( -38.60) >DroYak_CAF1 142125 119 - 1 UGUU-UUUGCAUGCAUGACAAUGUGUGCAGAAUACGCGCGCGCUCACACACUAAAACACUCGCACAACCGAAGGCGUGUGUGCAUGUAUGCAAUUUUUUGGGGUGGUAUUGGCGAAGGAA .(((-(((((((((((....)))))))))))).)).(((.((....(((.(((((((((.(((.(.......)))).))))(((....))).....))))).)))....)))))...... ( -34.90) >consensus UGUU_UUUGCAUGCAUGACAAUGUGUGCAGAAUACGCGCGCGCUCACACACUAAACCACUCGCACAACCGAAGGCGUGUGUGCAUGUAUGCAAUUUUUUGGGGUGGUAUUGGCGGUGAAG ......(((((((((((.....((((((.......))))))...(((((((.....(..(((......)))..).))))))))))))))))))........................... (-32.96 = -32.96 + 0.00)


| Location | 9,510,919 – 9,511,039 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.55 |
| Mean single sequence MFE | -39.94 |
| Consensus MFE | -32.89 |
| Energy contribution | -33.13 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
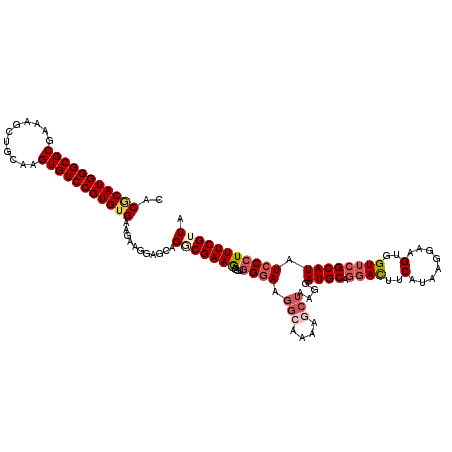
>3R_DroMel_CAF1 9510919 120 - 27905053 UCUUCUUACUUUCGCCCCCAGCAAUGAAACGCCAUGAAUUGGGGGAGACGGAAUCUAACGAUUGCGAACGCGUCGUAUCUUGUUUUUUGCAUGCAUGACAAUGUGUGCAGAAUACGCGCG ...........((.(((((((..(((......)))...))))))).))((.((((....)))).))..((((.(((((.......(((((((((((....)))))))))))))))))))) ( -44.31) >DroSec_CAF1 140597 119 - 1 UCUUCUUACUUUCGUCUCCAGCAAUGAAACUCCAUGAAUUGGGGGAGACUGAAUCUAACGAUUGCGAACGCGUUGUAUCUUGUU-UUUGCAUGCAUGACAAUGUGUGCAGAAUACGCGCG ..........((((((((((....))...(((((.....))))))))))..((((....))))..)))(((((.((((......-(((((((((((....)))))))))))))))))))) ( -37.40) >DroSim_CAF1 130758 119 - 1 UCUUCUUACUUUCGUCCCCAGCAAUGAAACGCCAUGAAUUGGGGGAGACGGAAUCUAACGAUUGCGAACGCGUUGUAUCUUGUU-UUUGCAUGCAUGACAAUGUGUGCAGAAUACGCGCG ...........((..((((((..(((......)))...))))))..))((.((((....)))).))..(((((.((((......-(((((((((((....)))))))))))))))))))) ( -40.50) >DroEre_CAF1 148446 119 - 1 UCUUCUUACUUUCUUCCCCAGCAAUGAAAAGCAAGAAAUUUGGGGAGAAGGAAUCAAACGAUUGCGAACGCGUCGUAUCUUCUU-UUUGCAUGCAUGACAAUGUGUGCAGAAUACGCGCG ..(((...((((((.(((((((........))((....)))))))))))))((((....))))..)))((((.(((((......-(((((((((((....)))))))))))))))))))) ( -38.90) >DroYak_CAF1 142205 119 - 1 UCUUCUUACUUUCUUUCCCAGCAUUGAAACGUAAAGAAUUGGGGGAGAAGGAAUCAAACGAGUGCGAACGCGUCGUAUCUUGUU-UUUGCAUGCAUGACAAUGUGUGCAGAAUACGCGCG ........((((((..(((((..((........))...)))))..)))))).................((((.(((((......-(((((((((((....)))))))))))))))))))) ( -38.60) >consensus UCUUCUUACUUUCGUCCCCAGCAAUGAAACGCCAUGAAUUGGGGGAGACGGAAUCUAACGAUUGCGAACGCGUCGUAUCUUGUU_UUUGCAUGCAUGACAAUGUGUGCAGAAUACGCGCG ..(((......((.(((((((..(((......)))...))))))).)).(.((((....)))).))))((((.(((((.......(((((((((((....)))))))))))))))))))) (-32.89 = -33.13 + 0.24)


| Location | 9,511,039 – 9,511,159 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.75 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -27.56 |
| Energy contribution | -29.16 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9511039 120 - 27905053 CACGCAUGGGCGCGAACGCUGCAAGUGUCCGUGUGAAGAAGGAGAAGGCGAAGAGGUGGGAAUCCAAAAGCUAAGGAUGCAGGACUUCAUUAGGAAGUGGUUCGCAUAUCCCUUUCGUUA ..(((((((((((...........)))))))))))...........(((((((.((((.(((((.....((.......))...(((((.....)))))))))).))...)).))))))). ( -37.10) >DroSec_CAF1 140716 120 - 1 CACGCAUGGGCGCGAAAGCUGCAAGUGUCCGUGUGAAGAAGGAGCAGGCGAAGAGGUGGGAAGGCAAAAGCUAAGGAUGCAGGACUUCAUAAGGAAGUGGUUCGCAUAUCCCUUUCGUUA ..(((((((((((...........)))))))))))...........(((((((....((((.(((....)))....((((.(((((.(........).))))))))).))))))))))). ( -37.50) >DroSim_CAF1 130877 120 - 1 CACGCAUGGGCGCGAAAGCUGCAAGUGUCCGUGUGAAGAAGGAGCAGACGAAGAGGUGGGAAGGCAAAAGCUAAGGAUGCAGGACUUCAUAAGGAAGUGGUUCGCAUAUCCCUUUCGUUA ..(((((((((((...........)))))))))))...........(((((((....((((.(((....)))....((((.(((((.(........).))))))))).))))))))))). ( -38.00) >DroEre_CAF1 148565 120 - 1 CACCCAUGGGCGCGAAAGCUGCAAGUGUCCGUGAGAAAAAGGAGCAGGCGAAAAGGUGGGAAGGGAUAAGCUAAGGAUGCAGGACUCCAUAAGAAAGCGGUUCGCAUAUGCCUUUCGAUA (((((((((((((...........)))))))))..........(....).....))))((((((.(((.((...(((........)))....(((.....))))).))).)))))).... ( -32.70) >DroYak_CAF1 142324 120 - 1 GACACAUGGGCGCGAAAGCUGCAAGUGUCCGUGUGAAGAAGGAGCAGACGAAAAGUUUGGAAGGGAUAAGCUAAGGAUGCACCAUUUCACAAGGAAGCGGUUCGCAUAUCCCUUUCGAUA ..(((((((((((...........))))))))))).....................((((((((((((.((.......))(((.((((.....)))).))).....)))))))))))).. ( -39.20) >consensus CACGCAUGGGCGCGAAAGCUGCAAGUGUCCGUGUGAAGAAGGAGCAGGCGAAGAGGUGGGAAGGCAAAAGCUAAGGAUGCAGGACUUCAUAAGGAAGUGGUUCGCAUAUCCCUUUCGUUA ..(((((((((((...........)))))))))))...........(((((((....((((.(((....)))....((((.((((..(........)..)))))))).))))))))))). (-27.56 = -29.16 + 1.60)

| Location | 9,511,079 – 9,511,199 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -42.19 |
| Consensus MFE | -34.80 |
| Energy contribution | -35.84 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 9511079 120 - 27905053 GGAGUCUCGCUGCUCUCUUCCUGUUCUUCGCACACAUACACACGCAUGGGCGCGAACGCUGCAAGUGUCCGUGUGAAGAAGGAGAAGGCGAAGAGGUGGGAAUCCAAAAGCUAAGGAUGC (((.(((((((..(((((((.(.((((((((((....((((..(((...(((....))))))..))))..)))))))))).).))))).))...))))))).)))............... ( -43.20) >DroSec_CAF1 140756 120 - 1 GGAGUCUCGCUGCUCUCUUCCUGUUCUUCGCACACAUACACACGCAUGGGCGCGAAAGCUGCAAGUGUCCGUGUGAAGAAGGAGCAGGCGAAGAGGUGGGAAGGCAAAAGCUAAGGAUGC ...((((..(..(.((((((((((((((..(((((..((((..((....))((....)).....))))..)))))....)))))))))..))))))..)..))))............... ( -43.90) >DroSim_CAF1 130917 120 - 1 GGAGUCUCGCUGCUCUCUUCCUGUUCUUCGCACACAUACACACGCAUGGGCGCGAAAGCUGCAAGUGUCCGUGUGAAGAAGGAGCAGACGAAGAGGUGGGAAGGCAAAAGCUAAGGAUGC ...((((..(..(.((((((((((((((..(((((..((((..((....))((....)).....))))..)))))....))))))))..)))))))..)..))))............... ( -43.50) >DroEre_CAF1 148605 120 - 1 GGAGUCUCGCUAUUCUCUUCCUGUUCUUCGCACACACACACACCCAUGGGCGCGAAAGCUGCAAGUGUCCGUGAGAAAAAGGAGCAGGCGAAAAGGUGGGAAGGGAUAAGCUAAGGAUGC ...((((.(((..((((((((..((.(((((............((((((((((...........))))))))).)............)))))..))..))))))))..)))...)))).. ( -38.25) >DroYak_CAF1 142364 120 - 1 GGAGUCUUGCUGCUCUCUUCCUGUUUUUCGCACACAUACAGACACAUGGGCGCGAAAGCUGCAAGUGUCCGUGUGAAGAAGGAGCAGACGAAAAGUUUGGAAGGGAUAAGCUAAGGAUGC ...((((((((..((((((((..((((((((...(((((.(((((.((.(.((....))).)).))))).)))))..(......).).)))))))...))))))))..)))..))))).. ( -42.10) >consensus GGAGUCUCGCUGCUCUCUUCCUGUUCUUCGCACACAUACACACGCAUGGGCGCGAAAGCUGCAAGUGUCCGUGUGAAGAAGGAGCAGGCGAAGAGGUGGGAAGGCAAAAGCUAAGGAUGC ...((((.(((....((((((..((((((((...........(((((((((((...........)))))))))))............))))))).)..))))))....)))...)))).. (-34.80 = -35.84 + 1.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:13 2006